Genetic map of Britain goes on display
A genetic map of the British people has been produced by Oxford University researchers. It forms the centrepiece of their display at the Royal Society's free Summer Science Exhibition, which opens today.
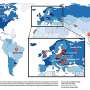
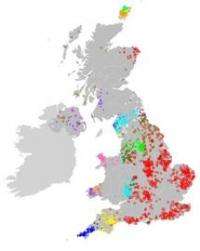
The remarkable thing about the map is how much people sharing similar gene variations cluster geographically.
The groupings often appear to match the separate historical pasts of different areas of Britain, following ancient enmities or reflecting differences that we hold onto today about where in the country we come from.
On the genetic map of Britain, Cornish people clustered separately from those from Devon, while the Scottish and Irish tended to share the same DNA markers. Those in South Wales formed a group, while there were separate clusters in the Welsh borders and in Anglesey in North Wales. People in Orkney were different from everyone else.
In England, the majority of the South, South-East and Midlands formed one large group. Cumbria, Northumberland and the Scottish Borders seemed to share a common past. And Lancashire and Yorkshire, despite their rivalry, seemed to be as one genetically.
'We first set out to map genetic variation across the UK,' says Professor Peter Donnelly, director of the Wellcome Trust Centre for Human Genetics and one of the scientists involved in the project. 'Our results show striking patterns of genetic clustering within different geographical regions of the UK. By comparing the different UK clusters with potential source populations from Europe we are able to learn more about the history of the people of the British Isles.'
The 'People of the British Isles' project began in 2004 with funding from the Wellcome Trust. Oxford University researchers travelled across the British Isles collecting blood samples from more than 4,000 people whose four grandparents all came from the same area.
The researchers used the latest DNA analysis techniques to look at how people's DNA sequence varied at 600,000 different sites across their whole genome.
They then grouped people together according to how much they tended to show the same small variations in their DNA sequence, and plotted them on a map of the British Isles. The analysis was done in collaboration with Dr Stephen Leslie, now at the Murdoch Childrens Research Institute in Melbourne, Australia.
Dr Bruce Winney of Oxford's Department of Oncology, a member of the research team, suggests you can form theories to explain the origins of the genetic variation the group has mapped out.
'We might be seeing the result of Anglo-Saxon invasions pushing other peoples down into Cornwall or Wales,' he suggests. 'Or we might be seeing how Britain was recolonised after the ice ages. The West of our islands may have been peopled by movement up the coastal areas from Atlantic-facing Europe, whilst the southeast was influenced by pre-Anglo-Saxon movements from the area that now spans Denmark to Belgium. These patterns may then have been reinforced by the Anglo-Saxon invasions much later.'
However, he cautions that these genetic differences that the project has found across the British Isles are small. We are far more genetically alike than we are different, he says.
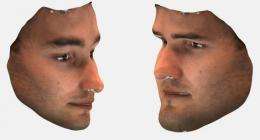
The Oxford University scientists now have further funding from the Wellcome Trust and are returning to the initial volunteers to collect digital 3D photographs of their faces.
Working in collaboration with Professor Josef Kittler and colleagues from the University of Surrey, the team will use the high-resolution photographs to measure particular features of a face, such as the size of the nose or mouth.
The aim is to understand some of the genetic basis behind the way we look. The researchers will look for links between our differing facial features and the variation in our DNA which they mapped out in the initial part of the project.
Professor Kittler explains: 'We measure the shape of facial features (nose, eyes) in 3D photographs of a large population of subjects and look for individuals at the extreme ranges, with a view to pinpointing those individuals with the largest genetic differences.'
Lead scientist Professor Sir Walter Bodmer, of the Department of Oncology at Oxford University, says: 'Everyone is interested in faces and why little Johnny has a mouth like his mother or a nose just like his father. We aim to find the genetic differences that explain that.
'Gene variants that determine facial features may be distributed differently in different countries, and perhaps even areas within a country, such as within the UK. We should be able to explain that too. Facial recognition, which enables us to recognise our kith and kin, has been very important in our evolution and we believe that unravelling the basis for that is a fascinating problem.'
Visitors to the researchers' stand at the Royal Society's Summer Science Exhibition this week will be able to see the genetic map of the British Isles, and find out how the map relates to the known archaeology and history of the country.
There's an opportunity to find out about the genes underlying our facial features. And the exhibit shows how our family names often indicate where in the country we come from, thanks to related research carried out at University College London under Professor Paul Longley. Visitors can view the distribution of their surname across Britain and the way that this has changed over the last 125 years.