Major new resource for fungal research
Researchers led by Conway Fellow, Prof Geraldine Butler have constructed a set of tools that will be of huge benefit to the scientific community as they try to understand the how the fungal pathogen, Candida parapsilosis causes infection.
Candida species are among the most common causes of fungal infection globally. C. parapsilosis causes infection outbreaks in neonatal wards and is one of the few Candida species that is transferred from the hands of healthcare workers.
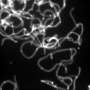
C. parapsilosis grows as biofilms or living mats of cells on the surface of indwelling medical devices such as feeding tubes. Until now, very little was known of the virulence properties of the pathogen and current treatment regimens involve removing the infected indwelling device.
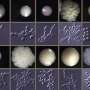
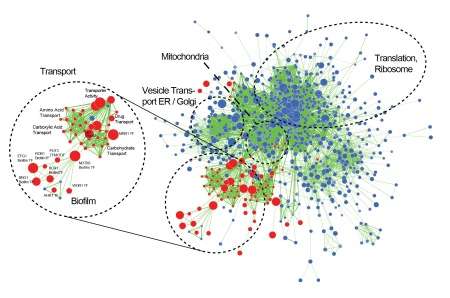
"Our aim in this study was to characterise the genes needed for biofilm development using a genetic approach. In order to find out how a gene works, a geneticist will often remove it or knock it out to see what happens as a result. We created a collection of 200 C. parapsilosis strains carrying knockouts in regulatory genes, and we identified those required for biofilm development", explained Prof Geraldine Butler.
The research team then looked at the impact on growth under various conditions such as temperature and the presence of anti-fungal drugs when particular regulatory genes are missing.
"We found similarities between C. parapsilosis and the most commonly isolated species, C. albicans but some regulators have major roles only in C. parapsilosis. Two particular transcription factors, Cph2 and Bcr1 are major biofilm regulators in C. parapsilosis only. Perhaps more importantly, we created a major research resource that will be accessible to the fungal research community globally", said Prof Butler.
More information: Holland, LM et al. "Comparative Phenotypic Analysis of the Major Fungal Pathogens Candida parapsilosis and Candida albicans." PLos Pathogens September 18, 2014. DOI: 10.1371/journal.ppat.1004365