Team publishes structural basis for HCV RNA replication
In this week's issue of Science, researchers at Gilead Sciences, Inc. and Beryllium reveal new details about how the hepatitis C virus (HCV) replicates its genome. HCV is estimated to affect 150-200 million people worldwide and is the major cause of liver transplantation in the US.
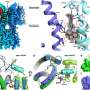
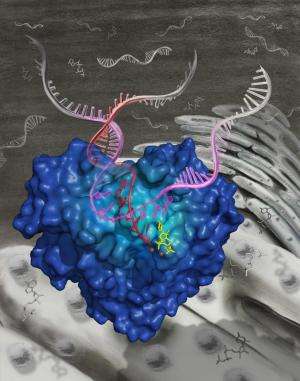
HCV uses RNA as genetic material which must be replicated in order to propagate the viral infection. Appleby et al. determined high resolution X-ray crystal structures of the HCV polymerase during the process of RNA replication, shedding light on the replication complex after 15 years of speculation. These molecular snap-shots unveil the NS5B catalytic mechanism in successive steps from the opening of the fist-like polymerase through the rapid RNA polymerization stage known as elongation. "For the first time we can see at the atomic level how the HCV polymerase interacts with the genomic RNA template, replicating RNA, and nucleotide substrates," said corresponding author Thomas Edwards of Beryllium.
Sofosbuvir acts during the elongation stage of HCV genomic replication, where the nucleotide triphosphate metabolite of the drug is incorporated into the growing RNA strand and terminates replication. The structural data presented by Appleby et al. demonstrate how the HCV polymerase recognizes sofosbuvir in a manner distinct from either native substrates or other nucleotide-based therapies.
"These structures advance our understanding of how an important member of the Flaviviridae family of viruses replicates genomic RNA," said William Lee, Senior Vice President of Research at Gilead Sciences. "This information will be useful in identifying replication inhibitors of other pathogenic viruses in this family responsible for human diseases."
More information: "Structural basis for RNA replication by the hepatitis C virus polymerase," by T.C. Appleby et al. Science, www.sciencemag.org/lookup/doi/ … 1126/science.1259210