It takes a village... to ward off dangerous infections? New microbiome research suggests so
Like a collection of ragtag villagers fighting off an invading army, the mix of bacteria that live in our guts may band together to keep dangerous infections from taking hold, new research suggests.
But some "villages" may succeed better than others at holding off the invasion, because of key differences in the kinds of bacteria that make up their feisty population, the team from the University of Michigan Medical School reports.
The researchers even show it may be possible to predict which collections of gut bacteria will resist invasion the best—opening the door to new ways of aiding them in their fight.
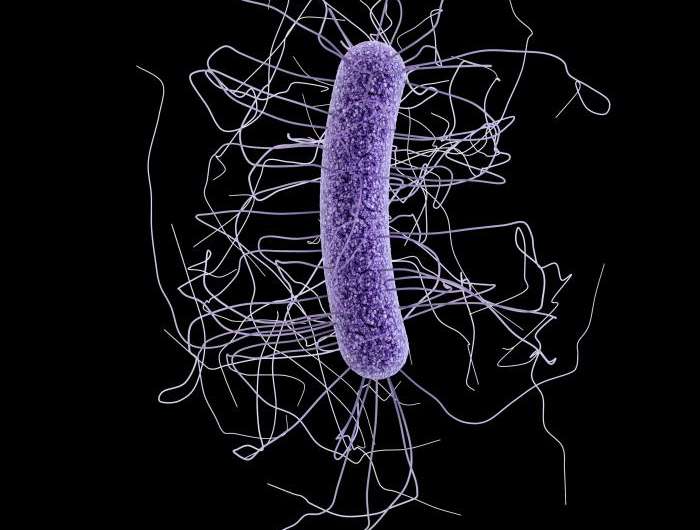
Working in mice, the team studied one of the most dangerous gut infections around: Clostridium difficile, which kills more than 14,000 Americans a year. C-diff also sickens hundreds of thousands more, mostly hospital patients whose natural collection of gut bacteria—their gut microbiome—has been disturbed by antibiotics prescribed to protect them from other infections.
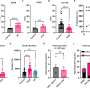
In a new paper published in the journal mBIO, the team reports the results from tests of seven groups of mice that were given different antibiotics, then were exposed to C-diff spores. The scientists used advanced genetic analysis to determine which bacteria survived the antibiotic challenge, and looked at what factors made it most likely that C-diff would succeed in its invasion.
The team also developed a computer model that accurately predicted C-diff's success rate for other mice in the study, based solely on knowing what bacteria the mice had in their natural gut 'village'. The model succeeded 90 percent of the time.
"We know that individual humans all have different collections of gut bacteria, that your internal 'village' is different from mine. But research has mostly focused on studying one collection at a time," says Patrick D. Schloss, Ph.D., the U-M associate professor of microbiology and immunology who led the team. "By looking at many types of microbiomes at once, we were able to tease out a subset of bacterial communities that appear to resist C-diff colonization, and predict to what extent they could prevent an infection."
He hopes that eventually, these models could serve as a diagnostic tool, to predict which patients will need the most protection against C-diff before they go to the hospital, or even to custom-design a protective dose of bacteria before or after a C-diff exposure.
In other words, to see which villages need the most reinforcements to prevail in battle.
Community matters
Schloss, who is a key member of the Medical School's Host Microbiome Initiative, notes that no one species of bacteria by itself protected against colonization. It was the mix that did it. And no one particular mix of specific bacteria was spectacularly better than others - several of the diverse "villages" resisted invasion.
Resistance was associated with members of the Porphyromonadaceae, Lachnospiraceae, Lactobacillus, Alistipes, and Turicibacter families of bacteria. Susceptibility to C. difficile, on the other hand, was associated with loss of these protective species and a rise in Escherichia or Streptococcus bacteria.
"It's the community that matters, and antibiotics screw it up," Schloss explains. Being able to use advance genetic tools to detect the DNA of dozens of different bacteria species, and tell how common or rare each one is in a particular gut, made this research possible.
Then, this massive amount of information about the villages of bacteria present in each of the mice in the experiment, and the relative success of each village in fighting off C-diff, was fed into the computer model created by the team.
The model uses a "random forest" statistical approach, a form of machine learning that allows the computer to weed through the data and make predictions based on what it learns. Schloss likens it to training an email spam filter to recognize and block scams from Nigerian imposters, based on a collection of keywords, while allowing emails from safe senders through.
Now that they have shown the concept in mice, the team is working to study the same issue in humans. They are already studying gut microbiome data from patients treated for symptoms of C. diff infection at the U-M Health System's hospitals, and will also study samples from patients with no C-diff symptoms to better understand the 'background' rate of how many of them carry the bug in a dormant, non-infectious state.
Although some patients at UMHS currently receive transplants of fecal material from healthy individuals to treat severe C-diff infection, Schloss notes that this treatment approach should likely only be a steppingstone to a more customized approach to re-arming the natural microbiome village inside vulnerable patients.
"It's an amazingly effective treatment, but if we could develop therapeutic probiotics instead, those would carry less risk," he says. "But first we have to know how the resistant communities are working."
More information: mBIO, DOI: 10.1128/mBio.00974-15, 14 July 2015 mBio vol. 6 no. 4 e00974-15 mbio.asm.org/content/6/4/e00974-15