Genetic sequencing offers same-day TB testing
Researchers have for the first time shown that standard tuberculosis (TB) diagnostic tests can be replaced by a sub-24 hour genetic test applied to the TB bacteria in a patient's sputum.
It currently takes up to two months to obtain the full diagnostic information for a patient with TB, as the bacteria grow very slowly in the laboratory. Scientists have sought for years to bypass this time-consuming step by examining the bacterial DNA directly from a sputum sample. However since most of the cells in sputum are human, it is difficult to spot the signal (TB DNA) within the noise (human and other bacteria) and even harder to find a method that might be affordable and practical across the world.
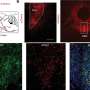
The new process, led by researchers from the University of Oxford and described in the Journal of Clinical Microbiology, rapidly processes the sputum to preferentially retain TB, using simple and relatively affordable materials, and then sequences and analyses the bacterial DNA. The Oxford team worked with researchers from the University of Nottingham, the Foundation for Medical Research, Mumbai, and Public Health England.
Until recently, DNA sequencing has required heavy machines and a well-equipped laboratory, which has limited its potential applications in the field. In this study, researchers have also shown that by using a new, real-time, handheld sequencing device (Oxford Nanopore MInION) they can achieve identical results, but with a process that might be applied anywhere in the world. In one example they achieved an effective turnaround time of 12.5 hours.
By using DNA sequencing, not only does this method detect drug-resistant TB bugs – vital information for the patient - but it also enables the tracking the geographical spread of strains, which is hugely valuable to public health workers, and something traditional tests cannot do.
TB is one of the top causes of death by infectious disease in the world, with 10.4 million cases of the disease in 2015, and 1.1 million deaths directly attributable to TB.
Dr Zamin Iqbal from the Wellcome Trust Centre of Human Genetics at Oxford University, who co-led the study, said: 'One of the great challenges with the management of TB is the need for rapid, comprehensive tests that do not require a hi-tech laboratory. We have shown that it is possible to get all information needed both for clinical management and for tracking disease spread, all within 24 hours of taking the sample from the patient. Further, by achieving this with a handheld device, we open the door to in-field diagnostic tests for TB.'
Dr Antonina Votintseva, lead author, said: 'Although genome sequencing has been used increasingly in research for analysing TB, the limiting factor has continued to be the weeks spent culturing the bacteria in the laboratory. By developing an affordable and simple method for extracting M. tuberculosis DNA direct from sputum, and thereby cutting turnaround time to below 24 hours, we have taken a great step towards comprehensive point-of-care diagnosis.
'There is more work to be done of course - our goal is to return test results before the patient leaves their clinic, with huge potential for reducing transmission of the disease, and of drug resistance.'
Dr Stephen Caddick, Wellcome Trust Director of Innovation, said: 'It can take many weeks for conventional tests for TB to provide results. Dr Iqbal and his team have made a significant breakthrough by developing a low-cost DNA extraction method which enables TB whole genome sequencing direct from patient samples and provides results in less than a day. The ability to use this technology to identify bacterial strains that may be resistant to antibiotic treatment, particularly in low and middle income countries, could be invaluable in the fight to tackle drug-resistant infections.'
More information: Antonina A. Votintseva et al. Same-day diagnostic and surveillance data for tuberculosis via whole genome sequencing of direct respiratory samples., Journal of Clinical Microbiology (2017). DOI: 10.1128/JCM.02483-16