New process could be key to understanding complex rearrangements in genome
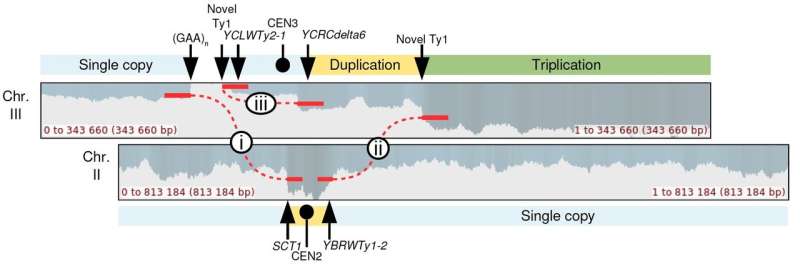
Understanding complex genomic rearrangements (CGRs), the culprit in the development of many types of cancer and genetic disorders, has always been a challenge because of the limitations of established DNA sequencing techniques. However, a team led by Tufts University biologists has successfully harnessed new technology to develop an approach that could allow for rapid and precise identification of the CGRs involved in disease, cancer and disorder development, which is critical for diagnosis and treatment. The results appeared this week in the December issue of Genome Research.
CGRs are created by DNA double-strand breaks that are not repaired properly. Many types of cancer are characterized by the presence of CGRs. They can also occur at the beginning of life, leading to severe congenital disorders such as Pelizaeus-Merzbacher disease. Repetitive DNA sequences, are also subject to double-strand breaks, which can lead to CGRs. Expansions of repetitive DNA sequences are implicated in hereditary diseases, such as Huntington's disease, fragile X syndrome and Friedreich's ataxia.
Mapping the outcomes of CGRs is crucial for understanding their causes and the potential impact they have on DNA and abnormal cellular formation. Ideally, this mapping would occur through high-resolution imaging. However, high-resolution bio-imaging of CGRs has traditionally been a laborious, time-intensive process that required a good deal of skill.
Led by Ryan J. McGinty, a Ph.D. candidate in the Department of Biology at Tufts University's Graduate School of Arts and Sciences, the research group sought to develop a more efficient process to analyze CGRs.
Use of a new hand-held DNA sequencing device was instrumental to the team's approach. Previous DNA sequencing technologies read DNA in segments of a few hundred nucleotides. The new device, developed by Oxford Nanopore Technologies, allowed researchers to analyze DNA fragments that were tens of thousands of nucleotides in length, showing a fuller picture of the CGRs.
Using a yeast model system, the researchers analyzed the longer fragments to study various CGRs resulting from unstable DNA repeats. Doing so enabled them to read complex events in their entirety and allowed them to uncover the underlying mechanisms that led to each of the complex events.
"This is a groundbreaking approach to analyzing CGRs and determining their origins and outcomes," said Sergei Mirkin, Ph.D., professor and chair of the Department of Biology at Tufts and the study's corresponding author. "This research could significantly advance the way the scientific community deciphers, diagnoses and treats certain genetic disorders and other diseases caused by disruptions of the genome."
The Mirkin Lab at Tufts studies DNA structure and functioning, with a primary focus on various DNA repeats, their role in the maintenance of the genome and their effects on major genetic transactions. A significant part of the lab's research is devoted to unusual DNA structures and their biological roles.
More information: Ryan J. McGinty et al, Nanopore sequencing of complex genomic rearrangements in yeast reveals mechanisms of repeat-mediated double-strand break repair, Genome Research (2017). DOI: 10.1101/gr.228148.117