Scientists develop low-cost way to build gene sequences
A new technique pioneered by UCLA researchers could enable scientists in any typical biochemistry laboratory to make their own gene sequences for only about $2 per gene. Researchers now generally buy gene sequences from commercial vendors for $50 to $100 per gene.
The approach, DropSynth, which is described in the January issue of the journal Science, makes it possible to produce thousands of genes at once. Scientists use gene sequences to screen for gene's roles in diseases and important biological processes.
"Our method gives any lab that wants the power to build its own DNA sequences," said Sriram Kosuri, a UCLA assistant professor of chemistry and biochemistry and senior author of the study. "This is the first time that, without a million dollars, an average lab can make 10,000 genes from scratch."
Increasingly, scientists studying a wide range of subjects in medicine—from antibiotic resistance to cancer—are conducting "high-throughput" experiments, meaning that they simultaneously screen hundreds or thousands of groups of cells. Analyzing large numbers of cells, each with slight differences in their DNA, for their ability to carry out a behavior or survive a drug treatment can reveal the importance of particular genes, or sections of genes, in those abilities.
Such experiments require not only large numbers of genes but also that those genes are sequenced. Over the past 10 years, advances in sequencing have enabled researchers to simultaneously determine the sequences of many strands of DNA. So the cost of sequencing has plummeted, even as the process of generating genes has remained comparatively slow and expensive.
"There's an ongoing need to develop new gene synthesis techniques," said Calin Plesa, a UCLA postdoctoral research fellow and co-first author of the paper. "The more DNA you can synthesize, the more hypotheses you can test."
The current methods for synthesizing genes, he said, either limit the length of a gene to about 200 base pairs—the sets of nucleotides that made up DNA—or are prohibitively expensive for most labs.
The new method involves isolating small sections of thousands of genes in tiny droplets of water suspended in an oil. Each section of DNA is assigned a molecular "bar code," which identifies the longer gene to which it belongs.
Then, the sections, which initially are present in only very small amounts, are copied many times to increase their number. Finally, small beads are used to sort the mixture of DNA fragments into the right combinations to make longer genes, and the sections are combined. The result is a mixture of thousands of the desired genes, which can be used in experiments.
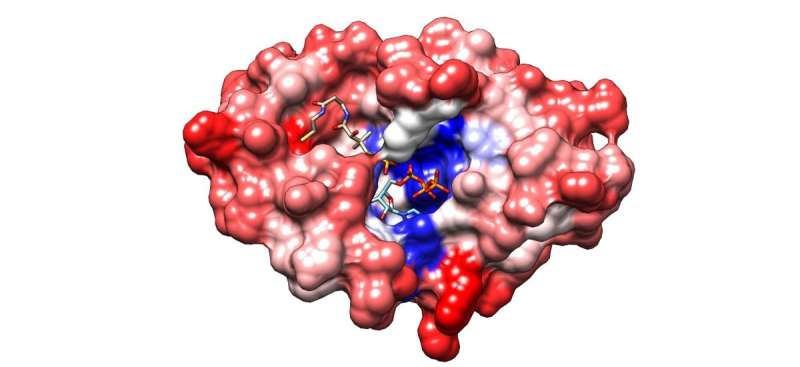
To show that technique worked, the scientists used DropSynth to make thousands of bacterial genes—each as long as 669 base pairs in length. Each gene encoded a different bacterium's version of the metabolic protein phosphopantetheine adenylyltransferase, or PPAT, which bacteria need to survive. Because PPAT is critical to bacteria that cause everything from sinus infections to pneumonia and food poisoning, it's being studied as a potential antibiotic target.
The researchers created a mixture of the thousands of versions of PPAT with DropSynth, and then added each gene to a version of E. coli that lacked PPAT and tested which ones allowed E. coli to survive. The surviving cells could then be used to screen potential antibiotics very quickly and at a low cost.
DropSynth could potentially also be useful in engineering new proteins. Currently, scientists can use computer programs to design proteins that meet certain parameters, such as the ability to bind to certain molecules, but DropSynth could offer researchers hundreds or even thousands of options from which to choose the proteins that best fit their needs.
The team is still working on reducing DropSynth's error rate. In the meantime, though, the scientists have made the instructions publicly available on their website. All of the chemical substances needed to replicate the approach are commercially available.
More information: Calin Plesa et al. Multiplexed gene synthesis in emulsions for exploring protein functional landscapes, Science (2018). DOI: 10.1126/science.aao5167