Research on mutation 'hotspots' in DNA could lead to new insights on cancer risks
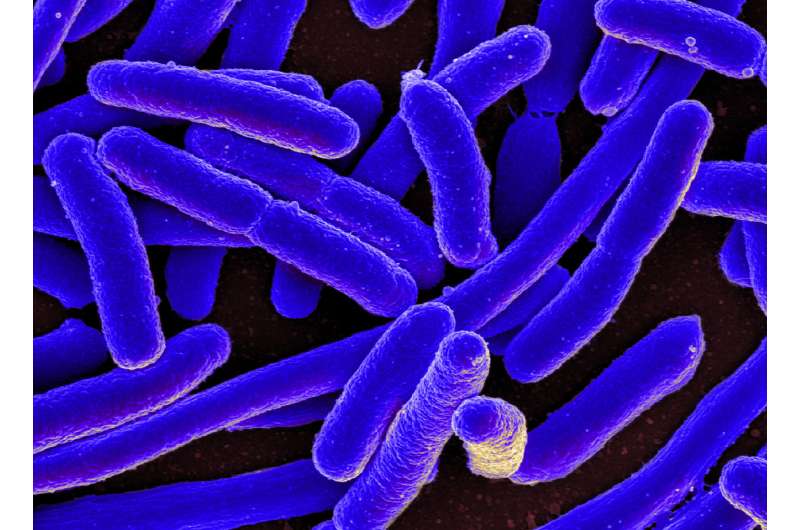
New research from Indiana University has identified "hotspots" in DNA where the risk for genetic mutations is significantly elevated.
These mutations arise because "typos" can occur as DNA replicates during cellular division. A recent analysis, which found that random mistakes in DNA play a large role in many cancer types, underscores the need to understand more about what triggers these errors.
The IU-led research, conducted in E. coli, appears in two papers in the "Highlights" section of the August issue of the journal Genetics. The "hotspots" identified are specific to E. coli and related bacterium, but the work could provide a roadmap to identifying similar trouble spots in human DNA.
"This research gets us closer to understanding how the cell's replication machinery interacts with DNA," said Patricia Foster, a professor emerita in the IU Bloomington College of Arts and Sciences' Department of Biology. "If you can understand exactly why an error occurs at a particular point on the DNA in bacteria, it gets you closer to understanding the general principles."
Foster is the first author on one of the two papers. The other paper's first author is Brittany Niccum, a Ph.D. student in Foster's lab at the time of the study.
The risk for cancer from DNA replication errors is highest in certain tissues—like the prostate and bones—where a higher rate of cellular renewal means there are more opportunities for mistakes to occur as the DNA is copied.
"There are parts of the genome that contain 'cancer drivers,' where changes in the DNA can allow tumor cells to proliferate," Foster said. "If you could know what sections of the DNA had a higher risk for mutation, you might be able to focus your analysis on these 'hotspots' to predict what will happen next."
In E. coli, the researchers found that the chances of DNA replication errors were up to 18 times more likely in DNA sequences where the same chemical "letter" in the sequence repeats multiple times in a row. They also found that errors were up to 12 times more likely in DNA sequences with a specific pattern of three letters.
These patterns of letters in the DNA sequence had been previously identified as common locations for replication errors. But Foster said the sheer volume of data in the new studies—with analysis across the bacteria's entire genome of 30,000 mutations accumulated during 250,000 generations—provide the "statistical weight" required to pinpoint the error rates with an unprecedented level of accuracy.
The studies also underline the importance of two systems in DNA replication: a "proofreader" enzyme and a molecular pathway called mismatch repair. Both serve as a defense against mistakes from the enzyme—called DNA polymerase—that copies the genome at a staggering rate of 1,000 letters per second.
This proofreader function resets the copying process after detecting a mistake. The IU researchers found that "switching off" this function caused 4,000 times more errors. Switching off mismatch repair, a backup system for the proofreader, caused 200 times more errors.
"When we switch off these backup systems, we start to see 'pure' errors—the places where the polymerase is more likely to make a mistake without intervention from other processes, " Foster said. "Until now, I don't think anyone could truly see the seriousness of these error hotspots in DNA."
More information: Patricia L. Foster et al, Determinants of Base-Pair Substitution Patterns Revealed by Whole-Genome Sequencing of DNA Mismatch Repair DefectiveEscherichia coli, Genetics (2018). DOI: 10.1534/genetics.118.301237
Brittany A. Niccum et al. The Spectrum of Replication Errors in the Absence of Error Correction Assayed Across the Whole Genome ofEscherichia coli, Genetics (2018). DOI: 10.1534/genetics.117.300515