Development and analytical validation of a next-generation sequencing based microsatellite instabili
The referenced article describes an assay that has a clinically relevant five-day turnaround time and can be conducted on as little as 20 ng genomic DNA with a batch size of up to forty samples in a single run.
Assay performance with respect to accuracy, reproducibility, precision, as well as control sample performance, was estimated across a wide range of FFPE samples of multiple histologies to address pre-analytical variability, and analytical variability.
Dr. Sean T. Glenn from OmniSeq Inc., Buffalo, NY 14203, USA, the Center for Personalized Medicine, Roswell Park Comprehensive Cancer Center, Buffalo, NY 14263, USA and the Department of Molecular and Cellular Biology, Roswell Park Comprehensive Cancer Center, Buffalo, NY 14263, USA said, "Microsatellite instability (MSI) is a well-described phenomenon characterized by the altered length of short repetitive regions of DNA referred to as microsatellites."
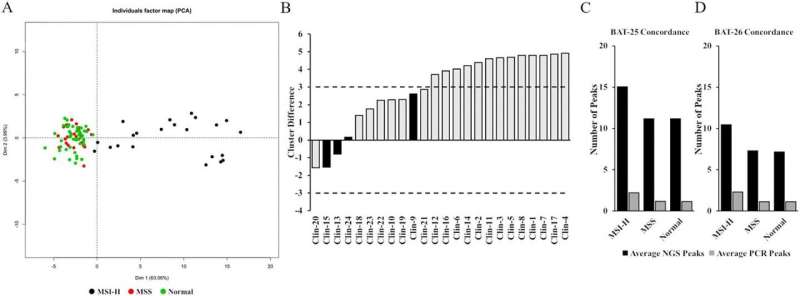
After amplification, fragment analysis chromatograms for each microsatellite are manually reviewed to assess differences between tumor and normal samples from the same patient in order to identify length differences, or so-called instability, in each microsatellite.
Although CRA and EEM account for the majority of microsatellite unstable tumors, microsatellite instability has a low but substantial incidence in various other tumors.
Consequently, the requirement of normal DNA can limit the number of patients that can have testing performed due to the difficulty to obtain normal tissue leading to suboptimal turnaround time or an inability to complete microsatellite testing.
Although there are additional published studies that use NGS testing to evaluate MSI status and those that use conventional fragment analysis without matched normal, the authors have developed the first agile NGS platform that is Clinical Laboratory Improvement Amendments certified and New York State Clinical Laboratory Evaluation Program approved for clinical MSI testing in patients using next-generation sequencing that can be utilized across all solid tumors without the need of matched normal tissue.
The Glenn Research team concluded, "Overall, we have developed the first NYS-CLEP approved, analytically validated MSI assay that requires minimum input of tumor DNA without the need for matched normal and is histology agnostic."
More information: Sarabjot Pabla et al, Development and analytical validation of a next-generation sequencing based microsatellite instability (MSI) assay, Oncotarget (2019). DOI: 10.18632/oncotarget.27142