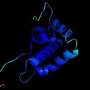
Building high-resolution protein models to fight COVID-19

During the COVID-19 pandemic, Michigan State University researchers are rising to the occasion and spearheading projects that seek to aid the international efforts to develop treatments for the virus. One such researcher is Dr. Michael Feig who is a part of the Department of Biochemistry and Molecular Biology at MSU. He and his postdoc Dr. Lim Heo generated high accuracy models for the SARS-CoV-2 virus. Seeing as these structural models were not available previously from experiment, these high accuracy models can now be used in further studies that test how certain chemicals respond to the proteins. These models are used as a starting point by other researchers for screening existing drugs or developing new drugs for their potential usage in treating COVID-19. The idea is that if the molecular structures of the drugs can bind to the SARS-CoV-2 virus, they can hinder its function and reduce the severity of COVID-19.
Within the greater effort to mitigate the impact of the virus on the population, there are three primary efforts. The first is vaccine development, the second is epidemiological modeling, and the third is to "find therapies either by repurposing existing drugs and eventually developing new drugs" according to Dr. Feig. Dr. Feig is working to reach the third goal. While other researchers with this goal are engaging in a clinical "trial and error" method, Dr. Feig's work enables a more precise approach that relies less on guesswork as it deals with the modeling and eventual testing of chemical reactions based on the modeled protein structures. His group specializes in high accuracy refinement and modeling of molecular structures where no experimental data is available and his group's work is particularly essential in that it provides the best models that can be generated today for SARS-CoV-2 proteins with unknown structures so that drug screening by other research groups can be more effective.
Of course, his work is also quite challenging. Though his group already has an established protocol for generating high accuracy models, Dr. Feig's group typically focuses on smaller protein targets. He stated that "because the targets in SARS-CoV-2 are quite large, they present computational challenges." Additionally he stated that "some of them are bound to membranes which also adds additional complexity that we had to address." Namely, the process his group uses is very computationally expensive and the larger and more complex the structure, the more computational resources are required to generate models for that structure. Not only do these complex structures need to be modeled, but they need to be modeled as quickly as possible due to the urgency of the COVID-19 pandemic. Dr. Feig said he had to "really push to get lots of models at a very high level of accuracy out." His group was able to overcome this challenge by acquiring as many computational resources at different sites as they can.
MSU's HPCC services have played a big role in helping Dr. Feig's research group overcome these difficulties. His group uses high performance computing (HPC) for the "computational challenge of using computer simulations to improve the models." In order to acquire the necessary tools to complete the models, Dr. Feig's group needs a high number of computer nodes. Their research requires an especially large number of GPUs. An allocation at national supercomputing centers helped address some of the need, but Dr. Feig expressed gratitude to ICER that it was "generous in giving us access to some of their GPUs" to provide significant additional resources to generate high accuracy models at a faster rate. The contribution of ICER's HPCC services allowed Dr. Feig's group to significantly accelerate the process through which they generate models.
Now, Dr. Feig has nearly finished generating high accuracy models for individual proteins of SARS-CoV-2 to send out to other research groups. Most generated models for the proteins are available available publicly and are starting to be used by other research groups who are working towards developing treatments for COVID-19. Dr. Feig explained that the next step for his group is to work on assembling those proteins into "more biologically relevant units" and eventually build a complete high accuracy model of the virus. This ongoing work is in collaboration with Professors Rommie Amaro at UCSD and Syma Khalid at the University of Southampton in the UK and aims at generating models for the viral envelope at high resolution. Such models could then give more insight into how the virus functions as a whole. One current focus is on assemblies that pertain to the spike protein. The spike protein is protein that facilitates the entry of the virus into the cell during the beginning stages of infection. By learning the mechanism by which the infection begins, it is easier for other groups to devise solutions that halt the infection process.
Prior to the pandemic, Dr. Feig already had a longstanding interest in modeling high resolution molecular structures. His work had historically dealt with building high accuracy models and improving methods for building high accuracy models. He said that before the pandemic he "didn't really think about coronaviruses but COVID-19 came along and there was a great need to have accurate models." Consequently, his group dropped everything once the pandemic began and "pushed really hard to generate models as good as we can to help the community and allow other researchers to be more successful."