New approach eliminates specific strains of a bacterium related to acne
A new study by the Translational Synthetic Biology research group at UPF presents a new approach to eliminating specific strains of a bacterium related to acne. The paper, published in the journal PLOS Pathogens, has also involved scientists from the company S-Biomedic and the University of Lund, in Sweden.
Our microbiome is made up of the microorganisms that live inside and outside our body. This complex microbial community primarily inhabits the skin, the oral mucosa and the gastrointestinal tracts, it is different in each of us and we live in symbiosis with it. Specifically, the skin microbiome is made up of multiple organisms such as bacteria, viruses and fungi. They live in equilibrium and some skin diseases, such as acne vulgaris, are associated with alterations to them.
The bacterium Cutibacterium acnes (C. acnes) is the most abundant in human skin. There are different strains of this bacterium, some predominate in healthy skin and others are associated with acne, which is a multifactorial disease. In healthy skin there is a balance and in acne there is a change in the abundance of certain strains, which brings about an imbalance known as dysbiosis.
Therefore, the use of antibiotic treatments is not optimal, as they usually kill the different strains of C. acnes and even other skin bacteria, thus altering the equilibrium. To address this problem, the team of researchers has tried a new approach that consists of manipulating the microbiome to achieve a potential therapeutic strategy to treat acne without affecting its equilibrium.
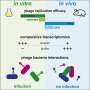
Through bacteriophage therapy it is possible to modulate the composition of C. acnes strains over time. We can reduce the strains associated with acne without affecting the ones that have beneficial features.
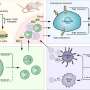
The strategy is based on one of the organisms found in the skin's microbiome: bacteriophages. These are viruses that infect bacteria and can help regulate them. "In our study we demonstrated that through bacteriophage therapy it is possible to modulate the composition of C. acnes strains over time. We can reduce the strains associated with acne without affecting the ones that have beneficial features," explains Marc Güell, study coordinator.
To address these strains specifically through bacteriophages, the scientists resorted to a mechanism that precisely bacteria have to prevent infections. They introduce modifications to their DNA that allow them to differentiate their own genetic material from that of others. "Using specific bacteriophages we attack pathogenic strains, which are the ones that do not have this defense strategy. The beneficial strains do have this defensive system against bacteriophages, so they are protected against infection," explains Nastassia Knödlseder, first author of the article.
As for future applications, Knödlseder explains that "we could, for example, use bacteriophages to 'clean' some of the existing strains that inhabit the skin. This would allow us to have more space available to better incorporate new bacteria and for them to remain."
"This work can help us to modulate the microbiome more efficiently, both to eliminate unwanted strains and to facilitate the introduction of new therapies," Güell concludes.
More information: Nastassia Knödlseder et al, Engineering selectivity of Cutibacterium acnes phages by epigenetic imprinting, PLOS Pathogens (2022). DOI: 10.1371/journal.ppat.1010420