Recommendations for in silico approaches for validating next-generation molecular sequencing analysis pipelines
The Association for Molecular Pathology (AMP), a global, molecular diagnostic professional society, today published consensus recommendations for the use of in silico approaches for validating Next-Generation Sequencing (NGS) data analysis pipelines.
The manuscript, "Recommendations for the Use of In Silico Approaches for Next Generation Sequencing Bioinformatic Pipeline Validation: A Joint Report of the Association for Molecular Pathology, Association for Pathology Informatics, and College of American Pathologists," was released online ahead of publication in The Journal of Molecular Diagnostics.
"As more laboratories around the country use in silico data to simulate variants to help validate the performance of clinical NGS data analysis pipelines, clinical laboratory professionals may need an aid for understanding both the value these methods bring and the important nuances and limitations of these approaches," said Justin Zook, Ph.D., Co-Chair of the AMP In Silico Pipeline Validation Working Group and Co-Leader of the Biomarker and Genomic Sciences Group at the National Institute of Standards and Technology.
"AMP convened a panel of subject matter experts from AMP, Association for Pathology Informatics, and College of American Pathologists to explore the advantages and disadvantages of these various types of in silico data. This new JMD report summarizes our key findings and provides useful recommendations to help clinical laboratory professionals select the most appropriate format for their specific purpose."
AMP devotes significant resources to developing evidence-based and consensus-driven practice guidelines to help establish essential elements of high-quality test design, such as analytical and clinical validity. The In Silico Pipeline Validation Working Group led the collective effort to review the available published literature and analyze the different types of in silico data and how they can be used in the clinical molecular diagnostic laboratory.
The new report also presents survey data on current use cases and highlights potential future applications. The team offered the following expert consensus opinion general recommendations based on these findings.
Consensus Recommendations
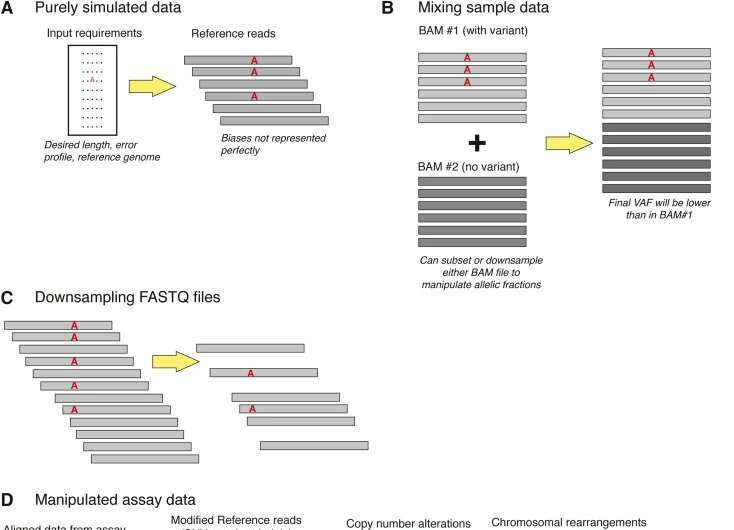
- The laboratory may use in silico data files to supplement NGS analytical validation, particularly to assess analytical sensitivity or false negative rates for specific variants; however, in silico data files cannot supplant the use of physical samples (e.g., patient samples).
- The laboratory should understand the functional limitations of the type(s) of in silico data being utilized.
- The laboratory should understand the limitations of most in silico data for assessing performance in particular genome contexts and variant types susceptible to systematic sequencing and mapping errors.
- The laboratory may consider using in silico samples for minor updates to clinical bioinformatics software pipelines.
- Commercial vendors and internal pipeline developers should include options in their analysis pipelines to facilitate easier in silico data file import and analysis by clinical laboratories.
"AMP is committed to collaborating with the broader clinical laboratory community to help identify areas of opportunity to continuously improve clinical practice, molecular diagnostic testing, and patient care," said Eric J. Duncavage, MD, Co-Chair of the AMP In Silico Pipeline Validation Working Group and Professor of Pathology and Immunology at the Washington University School of Medicine in St. Louis.
"This new joint report reviews, analyzes, and presents the latest data to help guide clinical laboratory professionals on when and how to use these important supplementary in silico data approaches. We intend to review and update these consensus recommendations as new data and other in silico methods become available."
More information: Eric J. Duncavage et al, Recommendations for the Use of In silico Approaches for Next Generation Sequencing Bioinformatic Pipeline Validation: A Joint Report of the Association for Molecular Pathology, Association for Pathology Informatics, and College of American Pathologists, The Journal of Molecular Diagnostics (2022). DOI: 10.1016/j.jmoldx.2022.09.007