This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
New tool to help harness human pangenome diversity for clinical interpretation of variants
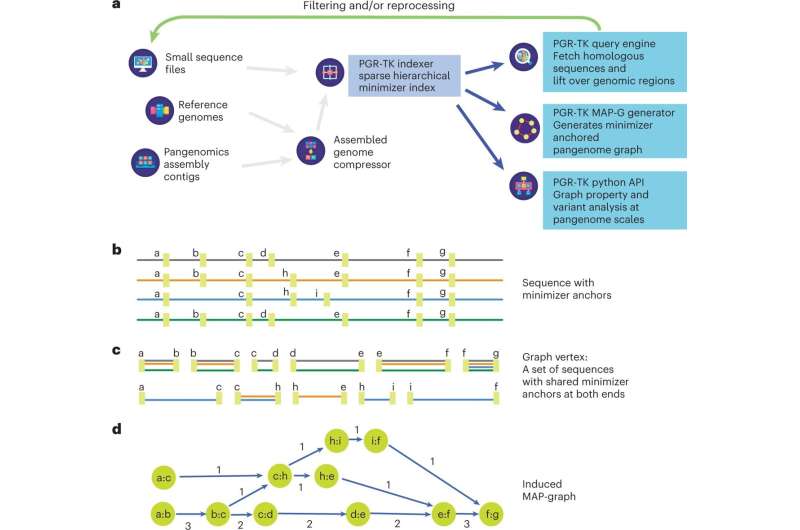
Researchers from GeneDx have published a paper titled "Multiscale analysis of pangenomes enables improved representation of genomic diversity for repetitive and clinically relevant genes" in Nature Methods. The work details research in which the researchers developed a new computational tool, the PanGenome Research-Tool Kit (PGR-TK), for scalable analysis of clinically relevant genes that were previously too complex to analyze.
"While new human pangenomes released last month by the Human Pangenome Reference Consortium inform a more comprehensive human genome reference, advanced tools are required to visualize the complex genetic structure and genetic variation across these diverse genome sequences," said Chen-Shan Chin, Ph.D., lead study author and Vice President, Genomic Technology and Algorithms at GeneDx. "The PGR-TK overcomes the limitations of other visualization graphs and helps resolve and visualize the most complex regions of the human genome that often affect multiple medical important phenotypes."
In analyzing the effectiveness of the PGR-TK, researchers applied the technology's graph decomposition methods to the Y-chromosome gene, DAZ1/DAZ2/DAZ3/DAZ4, of which structural variants have been linked to male infertility, and X-chromosome genes OPN1LW and OPN1MW, linked to eye disorders. The analysis showcased the ability of the PGR-TK to access and interpret 395 challenging but medically relevant genes previously inaccessible.
"The PGR-TK implements a novel algorithm that reveals fine structures in complex segmental duplications. It is the most comprehensive yet intuitive tool for studying these challenging regions and their possible association with phenotypic traits," said Heng Li, Ph.D., Associate Professor, Dana-Farber Cancer Institute and Harvard Medical School.
The PGR-TK is a software package with tools and algorithms to build an indexed sequence database that examines genetic sequences or regions with large-scale structural variations. This framework is scalable and enables analysis of multiple samples at varying resolution levels by adopting different parameters to facilitate exploratory analysis.
"I'm excited about new efficient methods to analyze and visualize the pangenome that can help researchers understand the causes of serious genetic diseases," said Justin Zook, Ph.D., co-author and Co-Leader of the Biomarker and Genomic Sciences Group at the National Institute of Standards and Technology (NIST). "They will also help us expand NIST's Genome in a Bottle benchmarks for understanding accuracy of methods in complex, medically relevant regions of the genome."
More information: Chen-Shan Chin et al, Multiscale analysis of pangenomes enables improved representation of genomic diversity for repetitive and clinically relevant genes, Nature Methods (2023). DOI: 10.1038/s41592-023-01914-y



















