This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
HuBMAP creates next generation of molecular analysis technologies and tools, enabling 3D tissue maps
More often than not, studies of human biology are conducted when the body is under duress from infection or disease. Now, as part of a larger effort to delineate what "healthy" looks like, two Stanford Medicine teams have unfurled detailed molecular maps of healthy human intestinal and placental tissues.
The maps, which capture cell types, cell quantity and other cellular nuances, are just two of a collection of maps that will establish a cellular baseline for the majority of the human body, including where cells in certain tissues congregate, how tissues develop during pregnancy and how cell-to-cell interactions drive human biology.
The studies, which will publish in Nature on July 19, are part of a larger effort spearheaded by the Human Biomolecular Atlas Program—called HuBMAP. It aims to fill gaps in our knowledge of how the human body works when it's in tip-top shape. Dozens of teams from the United States and Europe contribute to the HuBMAP consortium.
"In research, we have a habit of studying things that are abnormal without really understanding what normal looks like," said Michael Angelo, MD, Ph.D., an assistant professor of pathology who is also the co-chair of the HuBMAP steering committee. "That's created a big gap in our knowledge. HuBMAP is the only effort that is systematically focusing on the spatial architecture of these tissues."
By combining cellular imaging techniques, machine learning and other methods of molecular analyses, the teams are creating a comprehensive resource for researchers to better understand all human tissue. The data collected during the project, including imaging data and annotations, will be publicly available through HuBMAP. The goal is to enable researchers to study tissue-specific characteristics, understand disease mechanisms, and develop automated annotation tools that identify and characterize cells.
In contributing to mapping the human body overall, Michael Snyder, Ph.D., the Stanford W. Ascherman Professor of Genetics and former co-chair of the HuBMAP Steering Committee, and his team focused on the intestine, revealing key characteristics of this type of tissue:
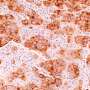
Cells in the intestine congregate in "neighborhoods," which are made up of different quantities of cell types with specific functions. The researchers were able to map these neighborhoods down to the level of individual cells.
The new maps revealed interesting clinical connections: The researchers found that tissue donors with higher body mass index had a greatly increased number of M1 macrophages, a type of immune cell associated with inflammation. And donors with a history of hypertension had fewer immune cells of a different type, called CD8 T cells, which play a role in seeking out and destroying possible cancer cells.
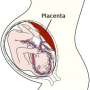
Angelo's team focused on the maternal-fetal tissue remodeling process, in which cells from the fetus embed into the uterine wall. Their study detailed several key pieces of developmental biology during pregnancy:
The team mapped the intricate, highly coordinated interaction between cells called trophoblasts, support cells that originate during early embryonic development, and the mother's immune system, which accommodates these unfamiliar cells as the placenta forms.
Corkscrew-shaped arteries that embed in the placenta and deliver blood and nutrients from the mom to the baby change their cellular make-up to ensure both cells from the baby and cells from the mother are incorporated into the placenta. This study was the first to sequentially show how these arteries change from un-remodeled to fully remodeled.
Snyder and Angleo hope that establishing detailed baselines, in coordination with dozens of other research teams creating cellular maps within HuBMAP, will help scientists further understand what happens when healthy bodies become diseased.
"These are really some of the first highly detailed spatial maps of the human body, allowing us to see not just what cells are there, but how they're organized," Snyder said. "It's hard to understand disease if you don't know what the healthy states look like. These maps will allow us to start comparing across different organs and analyzing what goes wrong during disease."
More information: Michael Snyder, Organization of the human intestine at single-cell resolution, Nature (2023). DOI: 10.1038/s41586-023-05915-x. www.nature.com/articles/s41586-023-05915-x
An atlas of healthy and injured cell states and niches in the human kidney, Nature (2023). DOI: 10.1038/s41586-023-05769-3. www.nature.com/articles/s41586-023-05769-3
A spatially resolved timeline of the human maternal–fetal interface, Nature (2023). DOI: 10.1038/s41586-023-06298-9. www.nature.com/articles/s41586-023-06298-9