This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Imaging breakthroughs provide insight into the dynamic architectures of HIV proteins
The human immunodeficiency virus (HIV) is a formidable pathogen. It mutates rapidly; in fact, estimates suggest that the genetic diversity of HIV in a single person at one point in time is equal to the diversity of influenza across the globe for a year. HIV also has developed structures to shield itself from recognition and attack by antibodies and therapeutics. These factors all contribute to HIV being a dangerous, hard-to-treat virus.
The more researchers can understand about the biological processes underlying how HIV infects cells, the better they can design treatments to penetrate the virus's defenses and destroy it. Now, Caltech researchers have imaged an elusive HIV protein structure at the atomic scale, seeing details with a resolution of one billionth of a meter.
The work was conducted in the laboratory of Pamela Björkman, David Baltimore, Professor of Biology and Biological Engineering and Merkin Institute Professor. A paper describing the study appears in the journal Nature. The study's first authors are Caltech postdoctoral scholars Kim-Marie Dam and Chengcheng Fan.
HIV primarily attacks immune cells called T cells, disabling them so that they cannot defend other cells in the body from infection. When an HIV virion prepares to enter a T cell, it undergoes some shape-shifting changes. These take place on the virus's so-called envelope protein, the protein on the surface of the virus that enables it to enter cells. As envelope proteins are so important to the virus's infection process, they are good targets for therapeutics or vaccines.
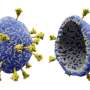
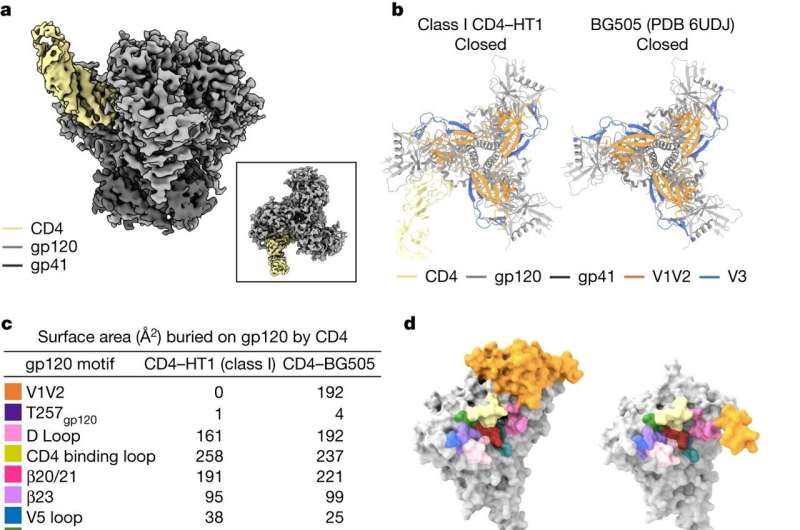
The HIV envelope protein is "trimeric," resembling a tripod-like flower with three "stem" portions—each called gp41—and three "petal" regions called gp120. To initiate the infection, each of the three gp120 proteins grabs on to a kind of receptor on the T cell called CD4. Once three CD4 receptors are secured by the three gp120 proteins, they expose sites that are recognized by a host co-receptor, and then a needle-like structure emerges from the stem regions of the "flower," enabling the virus to infect and gain entry into the human cell.
But what if the envelope protein's gp120 "petals" are only able to grab onto one or two CD4 receptors? Can the envelope protein still fully open up so the virus can infect the cell? Understanding this process could have significant implications for designing therapeutics. If researchers could prevent just one or two CD4 receptors from being grabbed by a gp120, would that be sufficient to thwart infection? To answer this open question, the team sought to image the envelope protein in these scenarios with only one or two CD4s bound.
"Structurally characterizing the intermediate envelope conformations is incredibly valuable to understand how HIV proteins work on a fundamental level," says Dam.
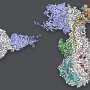
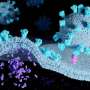
But imaging these structures is a challenge: Creating "heterotrimers," or envelope proteins that bind only one or two CD4 receptors, is not easy to do in a test tube for biochemical reasons. Through an innovative engineering approach, the team was able to devise a protocol to create stable heterotrimers. Then, utilizing Fan's expertise in a delicate procedure called cryo-electron microscopy, they were able to take images of the fragile heterotrimers' structures bound to CD4 receptors.
The structures showed that if only one or two CD4 receptors are bound, the envelope protein is not able to fully open up and undergo the shape-shifting process associated with infection.
"One major question that comes out of this work is: Can envelope proteins that do not fully open up still facilitate infection?" says Dam.
The team then shared results with the laboratory of Walther Mothes at Yale University, which was conducting similar attempts to image heterotrimers. Sharing information between the two labs showed that the behavior of engineered heterotrimers free-floating in a test tube (an experimental setup where proteins are in solution or soluble as opposed to bound to viral membranes) is remarkably similar to how envelope proteins on the viral surface behave in a more "real world" infection scenario.
This is an important finding because soluble constructs are used as the basis of developing new therapeutics, and it is critical to know if they are accurately mimicking natural processes.
Structural biology research like this is not only important for studying HIV but many different kinds of viruses.
"We've learned so much from HIV," says Dam. "When the COVID-19 pandemic started, we applied what we had learned from HIV to SARS-CoV-2."
"The structures of these previously unknown, intermediate envelope conformations offer fascinating insights into the structural changes driven by receptor interactions before host and viral membrane fusion," says Björkman. "Our research not only opens new avenues for exploring the complexities of HIV infection but also provides valuable insights that extend beyond therapeutic design, enhancing our overall understanding of viral dynamics."
More information: Dam, KM.A. et al, Intermediate conformations of CD4-bound HIV-1 Env heterotrimers, Nature (2023). DOI: 10.1038/s41586-023-06639-8. www.nature.com/articles/s41586-023-06639-8