This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
AI takes the reins in deep-tissue imaging
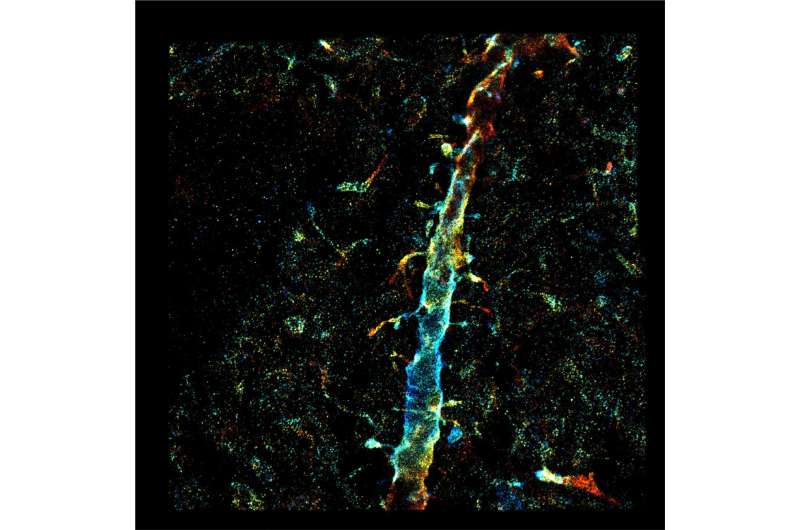
Artificial intelligence is finding more and more applications every day. One of the newest is in the biomedical field, using AI to control and drive single-molecule microscopy in ways no human can. The result is a landmark capacity of nanoscale optical imaging deep into tissue—visualizing 3D ultrastructure of the brain circuitry and plaque-forming amyloid beta fibrils in healthy and diseased brains, promising insights into autism and Alzheimer's disease.
Advances in deep-tissue imaging technology represent one of the most exciting developments in science today. They are allowing researchers to see more deeply and with greater resolution into the most elemental biological processes, shedding light on both human development and disease.
"Imaging through tissues is challenging, due to the distortion and blurring, called aberration, caused by the highly packed extracellular and intracellular constituents. In our imaging systems, which detect individual biomolecules in the three-dimensional tissue space with a precision down to a few nanometers, aberration is the disabling factor for visualizing molecular tissue architecture at its full definition," said Fang Huang, whose research team built the novel AI engine. Huang is the Reilly Associate Professor of Biomedical Engineering in the Weldon School of Biomedical Engineering at Purdue.
A patent-pending breakthrough was recently achieved through a multi-lab collaboration between the Huang Lab at the Weldon School; associate professor Alexander Chubykin's lab in Purdue's Department of Biological Sciences in the College of Science; and the lab of Gary Landreth, the Martin Professor of Alzheimer's Research at the Indiana University School of Medicine and Stark Neurosciences Research Institute.
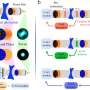
The investigators developed deep-learning-driven adaptive optics for their imaging system, which monitor the images of single-molecule emissions captured by the camera; measure the complicated aberrations induced by the tissue; and drive a complex, 140-element mirror device to compensate for and stabilize aberrations autonomously in real time.
The research was described in the journal Nature Methods, in a paper titled "Deep learning-driven adaptive optics for single-molecule localization microscopy."
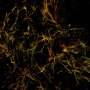
The research demonstrated deep-tissue, super-resolution imaging through 250 μm cut specimens (~1 order of magnitude improvement from previous demonstrations) at 20-70 nm resolution, resolving the ultrastructure of the dendritic spines and amyloid beta fibrils in the brain.
"The impact for biomedical science and medicine is huge: an opportunity to see more clearly and deeply into the recesses of compromised deep tissue in patients with autism or Alzheimer's Disease. Importantly, these advances in visualizing cellular anatomy will help to understand the pathophysiology of these conditions and inform novel treatment options in the future," said Chubykin, whose work focuses on understanding perceptual and learning impairments in autism through the functional and structural insights of the brain.
Optical microscopy enables visualization of cells and tissues up to a specificity level of details, namely the diffraction limit. Beyond the diffraction limit, however, smaller features in cells and tissues cannot be resolved. Traditional light microscopes widely available in universities, hospitals and high school classrooms all suffer from this fundamental limit of blurred vision of small objects—viruses, bacteria and small features inside cells or tissues can't be resolved.
Single-molecule localization microscopy has overcome this hurdle, enabling light-based observations with 10 to 100 times improved resolution. Now, with AI taking the reins of these "super-resolution" imaging systems, researchers are able to visualize the inner workings of cells and tissues at their full definition without hindrance.
"This technology arrived at a unique moment," said Landreth, whose lab focuses on the biological basis of Alzheimer's disease and how genetic risk factors influence disease pathogenesis.
"With the recent validation of therapeutic targeting of amyloid and preservation of cognitive function for Alzheimer's disease, our AI-driven, deep-tissue imaging system is positioned to advance the understanding of AD and evaluate potential therapeutics. This is a particular exciting time given the recent FDA approval of new AD drugs."
Huang disclosed the deep-learning-driven adaptive optics innovation to the Purdue Innovates Office of Technology Commercialization, which has applied for a patent with the U.S. Patent and Trademark Office to protect the intellectual property.
More information: Peiyi Zhang et al, Deep learning-driven adaptive optics for single-molecule localization microscopy, Nature Methods (2023). DOI: 10.1038/s41592-023-02029-0