This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Modeling viral evolution: A novel computational model with application to SARS-CoV-2 dynamics
Understanding the mutation and evolution of viruses (such as SARS-CoV-2) is crucial for effective public health management and response. Traditional epidemiological models often assume that viral transmissibility and pathogenicity remain constant during disease transmission, ignoring the fact that viruses continuously evolve through natural selection and random mutations. This simplification limits the accuracy of these models in predicting epidemic trends, especially when facing rapidly mutating viruses.
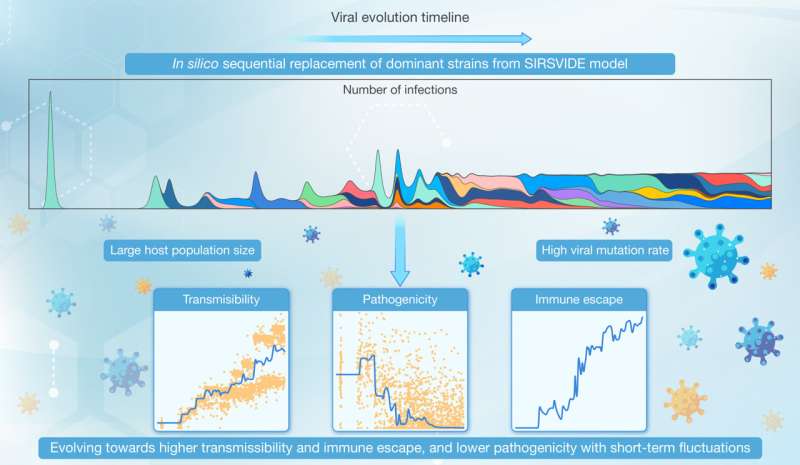
To overcome these limitations, Prof. Jian Lu's group at Peking University developed a novel computational model named SIRSVIDE (Susceptible-Infected-Recovered-Susceptible-Variation-Immune Decay-Immune Escape). The SIRSVIDE model not only incorporates basic principles of epidemiology but also integrates key features of viral mutation and evolution. The study is published in the journal hLife.
By simulating the dynamics of susceptible (S), infected (I), recovered (R) populations, and the process of individuals becoming susceptible again (S), while introducing elements such as viral variation (V), immune decay (ID), and immune escape (IE), the model can capture both short-term and long-term evolutionary dynamics of viruses. It considers not only the evolution of individual strains but also the competitive relationships among different strains, providing a universal framework for studying viral epidemiology and evolutionary dynamics.
Simulations under specific conditions (high mutation rate μ= 10-8, large host population N = 109) showed that viral populations undergo continuous lineage iterations and evolve towards increased transmissibility, enhanced immune escape, and reduced pathogenicity, accompanied by significant short-term fluctuations in viral traits. The study found that large host populations and high mutation rates are key factors leading to these unique evolutionary trends.
Despite these long-term evolutionary trends, the inherent randomness of viral evolution inevitably leads to short-term fluctuations in viral traits. Simulations under various parameters showed that a considerable proportion (27.12%-37.59%) of prevalent strains have both higher transmissibility and pathogenicity compared to their ancestral strains.
This suggests that new variants with simultaneously enhanced transmissibility and pathogenicity may emerge in the short term of an epidemic. Moreover, as the number of infections or mutation rate decreases, the uncertainty in the short-term evolutionary direction of viruses further increases.
The classical "transmission-virulence trade-off" hypothesis proposed by Andersen and May in 1982 suggests that there is a trade-off between transmissibility and pathogenicity in viral evolution. However, the transmission routes and pathogenic mechanisms of viruses are diverse, and transmissibility and pathogenicity are not always strictly coupled.
The SIRSVIDE model provides a dynamic analytical framework that comprehensively considers factors such as susceptible-infected-recovered-susceptible dynamics, immune decay, immune escape, and viral mutation, enabling in-depth analysis of the impact of different parameter changes on viral evolutionary dynamics. This helps us understand how viruses balance transmissibility and pathogenicity under multiple selection pressures to find the optimal adaptation strategy.
In summary, the SIRSVIDE model developed by Prof. Jian Lu's group provides a comprehensive framework for studying viral epidemiology and evolutionary dynamics. The simulation results reveal that under specific conditions, viral populations tend to evolve towards increased transmissibility, enhanced immune escape, and reduced pathogenicity, with large susceptible host populations and high mutation rates being key factors driving this evolutionary trend.
At the same time, the inherent randomness of viral evolution leads to short-term fluctuations in viral traits. These findings are consistent with the evolutionary evidence of SARS-CoV-2 and provide new insights for exploring the potential evolutionary patterns of other viruses, which is of great significance for guiding public health policy-making.
More information: Kaichun Jin et al, Modeling viral evolution: A novel SIRSVIDE framework with application to SARS-CoV-2 dynamics, hLife (2024). DOI: 10.1016/j.hlife.2024.03.006