Innate immune system proteins attack bacteria by triggering bacterial suicide mechanisms

A group of proteins that act as the body's built-in line of defense against invading bacteria use a molecular trick to induce bacteria to destroy themselves, researchers at the Indiana University School of Medicine have determined. The research could point the way toward new anti-bacterial treatments that could take on bacteria that are resistant to antibiotics.
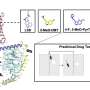
The proteins, called Peptidoglycan Recognition Proteins (PGRPs), are able to detect and target bacteria because bacteria are unique in having peptidoglycan polymers in their cellular walls. However, the mechanism by which PGRPs are able to kill bacteria had not been determined.
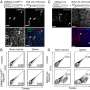
A research team led by Roman Dziarski, Ph.D., professor of microbiology and immunology at Indiana University School of Medicine – Northwest, reported May 22 in the advance online edition of the journal Nature Medicine that the PGRPs are able to induce a suicide response in the targeted bacteria.
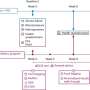
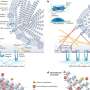
The PGRPs accomplish the mission by binding to specific sites in bacterial cell walls in ways that exploit a bacterial defense mechanism known as protein-sensing two-component systems. These systems, which normally enable the bacteria to detect and eject malformed proteins, interpret the PGRPs as just such malformed proteins. Unable to dislodge the PGRPs, the bacteria then activate a suicide response, the researchers said.
This approach is different than those employed by other anti-bacterial mechanisms, such as the immune system's white blood cells, said Dziarski.
"This could be a target to develop new anti-bacterial applications," Dziarski said.
Dziarski and colleague Dipika Gupta, Ph.D., associate professor of biochemistry and molecular biology at Indiana University School of Medicine – Northwest, first cloned the PGRP genes in 2001. The PGRP genes, which are found in species ranging from insects to mammals, are part of the body's innate immune system, in contrast to the mechanisms that learn and develop new immune responses to infections over time.
The PGRP proteins are normally expressed in phagocytic cells in blood and on body surface areas such as skin, mouth, intestine and other tissues that have direct or indirect contact with the external world, Dziarski noted. In some tissues it appears that the PGRPs help maintain a healthy relationship between the body and certain beneficial bacteria. Some studies have indicated that the loss of the PGRP proteins may lead to inflammatory bowel disease, suggesting that the research reported Monday could point the way to new approaches to target such problems, Dziarski said.