Genomic analysis of superbug provides clues to antibiotic resistance
An analysis of the genome of a superbug has yielded crucial, novel information that could aid efforts to counteract the bacterium's resistance to an antibiotic of last resort. The results of the research led by scientists from The University of Texas Health Science Center at Houston (UTHealth) are published in the Sept. 8 issue of the New England Journal of Medicine.
Superbugs are bacteria that are resistant to multiple antibiotics and represent one of the most challenging health problems of the 21st century. Infections caused by these bacteria can lead to longer illnesses, extended hospital stays and in some instances death. Antibiotic resistance is on the rise and alternative treatments are frequently suboptimal.
Researchers focused on a superbug called vancomycin-resistant enterococci (VRE), which is an intestinal bacterium that is resistant to multiple antibiotics, particularly vancomycin, a drug that has been used for treatment of potentially lethal hospital-associated infections.
"It is the second most common bacterium isolated from patients in U.S. hospitals after staphylococci," said Cesar Arias, M.D., Ph.D., the study's lead author and principal investigator. He is an associate professor of medicine at the UTHealth Medical School.
"The problem is that VRE has become so resistant that we don't have reliable antibiotics to treat it anymore," Arias said. "Daptomycin is one of the few antibiotics left with activity against VRE and is usually used as a drug of last resort. Additionally, this particular superbug is frequently seen in debilitated patients such as those in critical care units, receiving cancer treatment and patients receiving transplants, among others; therefore the emergence of resistance during therapy is a big issue."
VRE itself can develop resistance to daptomycin during treatment. To find out why, researchers compared the genomes of bacterial samples drawn from the blood of a patient with VRE bloodstream infection receiving daptomycin. The bacterium developed daptomycin resistance and the patient subsequently died.
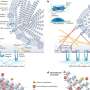
By comparing the genetic makeup of the bacterium before and after it developed resistance to daptomycin, the researchers were able to identify changes in genes directly tied to antibiotic resistance. "Our research provides direct substantiation that changes in two bacterial genes are sufficient for the development of daptomycin resistance in VRE during therapy," Arias said.
Barbara Murray, M.D., coauthor and director of the Division of Infectious Diseases at the UTHealth Medical School, said, "These results lay the foundation for understanding how bacteria may become resistant to daptomycin, which opens immense possibilities for targeting the functions encoded by these mutated genes. This would be a step toward the development of much needed new drugs. That is, once we understand the exact mechanism for resistance, one can start to develop strategies that block or attack the resistance mechanism."
Murray, holder of the J. Ralph Meadows Professorship in Internal Medicine, added, "This study identified genes never before linked to antibiotic resistance in enterococci. The genomic approach used in the study is very powerful and was able to pinpoint exactly the specific genes and mutations within them that resulted in the failure of daptomycin (CUBICIN®) therapy and contributed to the fatal outcome of the patient."
Arias' laboratory is doing additional research needed to determine the precise mechanisms by which the gene changes allow the bacterium to defeat the antibiotic. "There are mutations that appear to alter the bacterial cell envelope, which is the target of the antibiotic. The modifications brought about by the gene mutations may change the cell envelope to avoid the killing by these antibiotics. We believe these changes are a general mechanism by which bacteria protect themselves," Arias said.
Herbert DuPont, M.D., holder of the Mary W. Kelsey Distinguished Professorship in the Medical Sciences and director of the Center for Infectious Diseases at The University of Texas School of Public Health, said, "Twenty years ago antibiotic-resistant bacteria more often caused hospital-acquired infections in people with underlying illness or advanced age. Now, resistant bacteria are often seen in the community in otherwise healthy people, making treatment very complicated."
More information: The study is titled "Genetic Basis for In Vivo Daptomycin Resistance in Enterococci".