New study uncovers how brain cells degrade dangerous protein aggregates
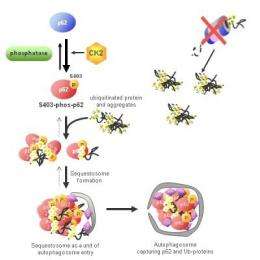
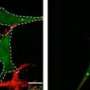
Researchers at the RIKEN Brain Science Institute (BSI) have discovered a key mechanism responsible for selectively degrading aggregates of ubiquitinated proteins from the cell. Their findings indicate that the capture and removal of such aggregates is mediated by the phosphorylation of a protein called p62, opening the door to new avenues for treating neurodegenerative diseases such as Huntington's disease and Alzheimer's disease.
One of the most important activities of a cell is the production of proteins, which play essential functions in everything from oxygen transport, to immune defense, to food digestion. Equally important to the cell's survival is how it deals with these proteins when they pass their expiry date: damaged or misfolded proteins have been associated with a range of debilitating conditions, including neurodegenerative diseases such as Alzheimer's disease.
In eukaryotic cells, the recycling of damaged or misformed proteins is governed by a small regulatory protein called ubiquitin in a process called "ubiquitination". By attaching itself to a protein, a ubiquitin molecule can tag the protein for destruction by proteasomes, large protein complexes that degrade and recycle unneeded proteins in the cell. This recycling of proteins by proteasomes is crucial to the maintenance of cellular homeostasis.
With their research, the BSI research group sought to shed light on one area where proteasome-based recycling falls short: protein complexes or aggregates, which proteasomes have trouble degrading. The group shows that this weakness is made up for by the phosphorylation of a protein called p62 at the serine 403 (S403) loci of its ubiquitin-associated (UBA) domain, which triggers a catabolic process called selective autophagy that degrades protein aggregates. It does this by forming a "sequestosome", a structure which sequesters polyubiquitinated protein aggregates in preparation for autophagy.
Published in the journal Molecular Cell, the discovery of this mechanism opens the door to the development of new, more effective drugs for selectively degrading protein aggregates, promising applications in the treatment of a range of neurodegenerative diseases.