New melanoma driver genes found in largest DNA sequencing study to date
(Medical Xpress) -- Yale Cancer Center geneticists, biochemists, and structural biologists have painted the most comprehensive picture yet of the molecular landscape of melanoma, a highly aggressive and often deadly skin cancer. The study appears in the July 29 advance online publication of Nature Genetics.
Melanoma, precipitated mainly by excessive exposure to the sun’s ultra-violet (UV) radiation, causes the vast majority of all deaths related to skin cancer. There will be around 76,000 new cases of melanoma and 9,000 deaths from the disease in the United States this year.
The Yale study used powerful DNA sequencing technologies to examine 147 melanomas originating from both sun-exposed and sun-shielded sites.
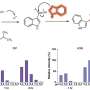
The study revealed an excess of UV-induced mutations in sun-exposed melanomas. Most of these are passenger mutations that do not have a functional role in melanoma. “We devised a mathematical model to sort out the relevant DNA alterations from over 25,000 total mutations,” says lead author Michael Krauthammer, associate professor of pathology, who directed the bioinformatics effort of the study.
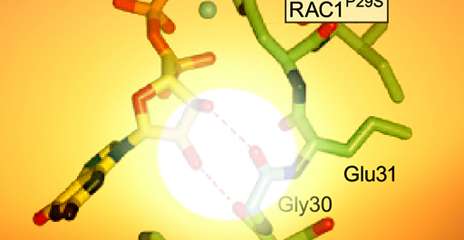
The analysis identified a frequent “gain-of-function” mutation in the RAC1 gene that has all the hallmarks of UV-damage. The study provided evidence that the mutant protein induces accelerated growth and movements among normal pigment cells, which are melanoma’s cells of origin. “It likely occurs at an early stage of tumor development and promotes malignant cell growth and spread to distant sites,” said corresponding author Ruth Halaban, senior research scientist at Yale School of Medicine and a member of Yale Cancer Center.
The Yale scientists say the RAC1 oncogenic mutation occurred in about 9% of melanomas from sun-exposed skin, and is the third most frequent mutation after the known BRAF and NRAS. They believe the prevalence of RAC1 mutation warrants development of therapies targeting that particular pathway.
The team also identified mutations that disable proteins — known as tumor suppressors — which suppress malignancy. Notably, the mutated protein known as PPP6C occurred only in tumors already mutated in BRAF and NRAS genes. “Our study mapped out a new, cooperative pathway for cancer development,” Halaban explained.
Finally, the study reveals new insights into the rarer melanomas from parts of the body shielded from the sun. Instead of mutations, these melanomas had duplicate copies of known oncogenes.