August 9, 2013 report
Study monitors DNA breaks and chromosome translocations in real time
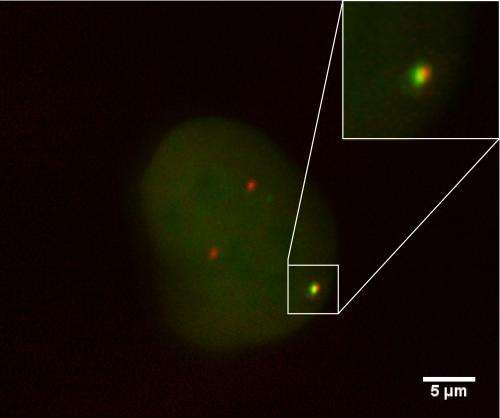
(Medical Xpress)—Researchers in the U.S. have developed a new method to study damage to DNA and resultant translocations in living cells.
DNA damage occurs regularly in living cells as a result of normal cellular processes and because of environmental factors such as radiation. The damage is constantly repaired, but if the repairs fail a break may occur in the two DNA strands and the two sections of the double helix then drift apart. This is referred to as a double-strand break (DSB), and is dangerous to the host cell because when the broken strands attempt to pair off again they have no template to follow and can pair with different chromosomes, producing a chromosome translocation, which is an unexpected rearrangement of the genes. Chromosome translocations are a hallmark of cancer cells.
Researcher Vassilis Roukos, of the National Cancer Institute in Bethesda, Maryland, and colleagues used ultra-high throughput time-lapse microscope imaging to monitor chromosome translocations in real time. The team's aim was to study the events during and after translocations in order to increase our understanding of these processes, and they succeeded in capturing images of DSBs and other translocation events as they occurred.
The team used living mouse cells with their DNA molecules engineered to split when exposed to a specific enzyme. They used fluorescent proteins to tag the broken ends and then used their microscope time-lapse imaging system to watch what happened over the next 36 hours.
They found that there were three distinct phases in chromosome translocation. The first was the DNA segments moving around a little, in an apparent random search for a "partner" for the broken strands to pair with; the second was for two broken segments to line up, and the third was for the segments to join together. In most instances, the DSB segments immediately joined up with the correct partner, but on rare occasions two DSBs of different chromosomes paired instead. Co-author Tom Misteli, also of the National Cancer Institute, said these translocations are rare, being seen in only one in around every 300 cells.
The researchers also discovered that chromosome translocations commonly occurred within hours of the double-strand breaking, and that their occurrence was unrelated to the cell cycle.
Another finding was that the cell's DNA repair system enzymes had an influence on the formation of translocations. For example, when the researchers disabled one of these enzymes, DNA-dependent protein kinase (DNAPK) in some of the cells, a chromosome translocation was nearly 10 times as likely to occur than in cells with active DNAPK. Misteli said that while DNAPK was previously known to play a role in translocation formation, little was understood about how it operates, and the new study was able to shed more light on the processes involved. One finding was that the incorrect strands still line up when DNAPK is disabled, but they are much less likely to join together, indicating that DNAPK is necessary to prevent incorrect pairings.
Misteli said the next step in the research is to try to find ways of preventing the DNA repairs from going wrong. The paper was published on 9 August in the journal Science.
More information: Spatial Dynamics of Chromosome Translocations in Living Cells, Science 9 August 2013: Vol. 341 no. 6146 pp. 660-664 DOI: 10.1126/science.1237150
ABSTRACT
Chromosome translocations are a hallmark of cancer cells. We have developed an experimental system to visualize the formation of translocations in living cells and apply it to characterize the spatial and dynamic properties of translocation formation. We demonstrate that translocations form within hours of the occurrence of double-strand breaks (DSBs) and that their formation is cell cycle–independent. Translocations form preferentially between prepositioned genome elements, and perturbation of key factors of the DNA repair machinery uncouples DSB pairing from translocation formation. These observations generate a spatiotemporal framework for the formation of translocations in living cells.
© 2013 Medical Xpress
















