A wrong molecular turn leads down the path to Type 2 diabetes
Computing resources at the U.S. Department of Energy's (DOE) Argonne National Laboratory have helped researchers better grasp how proteins misfold to create the tissue-damaging structures that lead to type 2 diabetes. The structures, called amyloid fibrils, are also implicated in neurodegenerative conditions such as Alzheimer's and Parkinson's, and in prion diseases like Creutzfeldt-Jacob and mad cow disease.
The results pinpoint a critical intermediate step in the chemical pathway that leads to amyloid fibril formation. With the new culprit in view, future work could target a possible treatment, such as designing an inhibitor to interfere with the harmful pathway. The results also helped reconcile earlier data from other labs that until now appeared contradictory.
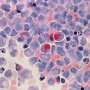
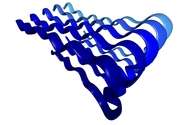
An amyloid fibril is a large structure consisting of misfolded proteins. Such fibrils form plaques, or areas of tissue damage, that researchers can observe with microscopes. Fibrils are believed to arise when proteins deviate from their normal 3D structures and instead adopt misfolded states that tend to clump together.
Like puzzle pieces, proteins are only useful when they have the correct shape. And since the fibrils they form when misfolded are strong, scientists believe that hope primarily lies not in dismantling them, but in heading off the folding errors.
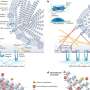
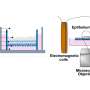
The researchers used two main approaches to identify the intermediate step and understand the pathway. University of Wisconsin-Madison professor Martin Zanni used a sophisticated technique that relies on 2-D infrared spectroscopy to follow the sequence of events in the chemical reactions leading to fibril formation. His technique can measure extremely fast processes using very small samples.
Then Juan de Pablo and Chi-Cheng Chiu from the University of Chicago's Institute for Molecular Engineering interpreted Zanni's measurements with data from molecular simulations to arrive at a complete picture of the early events leading to amyloid formation. De Pablo and Chiu used Intrepid, an IBM Blue Gene/P computer system at the Argonne Leadership Computing Facility (ALCF), and resources at the University of Chicago Research Computing Center. De Pablo and Chiu composed, ran and interpreted large-scale computer simulations of the pathway in action, and the results supplied an essential model of the molecular steps involved in the reaction.
"Using only one of the two methods would have been like running a race with only one leg," de Pablo said. "By combining both, computation and experiment, we can get to our answers faster and more dependably."
Together, researchers located an entire step that had been missing, and whose absence had been fueling confusion.
An earlier study indicated that the intermediate step was likely a floppy loop area formed by proteins, which didn't seem compatible with the tough, damaging fibril as an end result. Researchers believed that the fibrils should come from a rigid structure called a β-sheet.
The new data show, however, that both structures occur as the reaction changes over time. Transient rigid β-sheets form, then morph into floppy protein loops, which finally take the form of more β-sheets. The final β-sheets bind together and stack up to form the damaging fibrils.
The focus now will be to target the new intermediate step. With more data, researchers could design an inhibitor drug to bind to the offending protein, block the molecule and halt the pathway's progression.
Next, de Pablo intends to learn more about the particular protein intermediate that is implicated in type 2 diabetes. He has examined the basic units and small aggregates consisting of two or at most three molecules. "Now we need to understand how these small aggregates disrupt cell membranes," he said. "We also want to decipher how the fibril grows from a small nucleus."
To do so, he is pushing forward with plans to investigate bigger systems by using more supercomputing clout. He was recently awarded computing time on Argonne's IBM Blue Gene/Q, called Mira, the newest resource available to users at the ALCF. Mira is a 10-petaflops computer that packs 10 quadrillion calculations into each second of computing time.
De Pablo, Zanni and their collaborators will also apply the method from this publication to determine the intermediate steps in diseases other than type 2 diabetes, including neurodegenerative diseases like Alzheimer's. Scientists attribute more than 20 human diseases to the formation of amyloid fibrils. In each disease, the misfolding of a specific protein—a different one in every disease—is what triggers the problematic intermediate β-sheet.
"We want to understand the broader origins of the misfolding and aggregation problem," de Pablo said, "which we can do by looking at a wide range of molecules associated with different diseases. The eventual goal is to answer a few vital questions: What are the early stage misfolding events and small aggregates that form? How do they form? And how can we design inhibitors to stop them from forming?"
The findings are described in a paper published November 11 in the Proceedings of the National Academy of Sciences, titled "Mechanism of IAPP amyloid fibril formation involves an intermediate with a transient β-sheet." Also contributing to the research were scientists from the University of California-Irvine and the State University of New York at Stony Brook.
More information: www.pnas.org/content/early/201 … /1314481110.abstract