Early brain development implicated in Restless Legs Syndrome
In a study published online in Genome Research, researchers of the Helmholtz Zentrum München und the Technische Universität München have demonstrated that a common genetic variant associated with Restless Legs Syndrome (RLS) alters the expression of a critical gene during fetal development of the brain. This leads to alterations of the developing forebrain indicating an anatomical region involved in RLS.
Restless Legs Syndrome (RLS), a neurological disorder characterized by unpleasant sensations in the legs and the urge to move them, is not caused by a single genetic defect, but rather is a complex disorder influenced by many genetic and environmental components. Previously, researchers identified genetic variants in RLS patients; however, how these variants, each of which only has a small effect, contributed to RLS was unclear.
MEIS1 gene variant leads to altered development of the brain
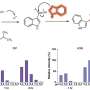
In this new study, authors from the Helmholtz Zentrum München (HMGU) and Technische Universität München as well as the Stanford Center for Sleep Medicine and Sciences demonstrate how one of these variants may contribute to RLS. The RLS-associated variant is located in a non-coding region of the MEIS1 gene and led to decreased ability to activate gene expression. Specifically, the authors observed the reduced gene expression in the future basal ganglia in the forebrain. "Here we have pinpointed down to an anatomical region for RLS," says lead author of the study, Prof. Juliane Winkelmann from HMGU, who is currently doing research at the Stanford University.
"The RLS-associated variant is located in an intron of MEIS1, a transcription factor involved in organ development and maintenance. The risk variant binds more strongly to the transcriptional regulator CREB1, which may lead to the reduced MEIS1 expression", explains Prof. Dr. Wolfgang Wurst from HMGU. Furthermore, screening analyses in animal models with reduced MEIS1 expression, conducted by the Institute of Experimental Genetics at the HMGU, led by Prof. Dr. Martin Hrabě de Angelis, showed hyperactivity, which resembles the human condition of RLS.
Reduced gene activity predisposition for RLS
Interestingly, the non-coding region only seems to be active during early brain development, suggesting that RLS, which is associated with aging, may have fetal origins. "Minor alterations in the developing forebrain during early embryonic development are probably leading to a predisposition to RLS", Winkelmann said. "Later in life, during aging, and together with environmental factors, these may lead to the manifestation of the disease."
In further studies researchers aim to investigate the affected cells in the forebrain. Based on their findings new treatment strategies for RLS may be developed.
This study provides one of the first in-depth examinations of a genetic variant identified in a genome-wide association study, which examines many individuals for genetic variants that are linked to a trait. Although many variants are often reported in these studies, it has been difficult understand how variants contribute to disease because they often lie in non-coding regions of the genome and have small effect sizes. This work also reveals that combinatorial use of multiple approaches will be likely required to unravel the physiological causes most of human diseases.
More information: D. Spieler, M. Kaffe, F. Knauf, J. Bessa, J. Tena, F. Giesert, B. Schormair, E. Tilch, H. Lee, M. Horsch, D. Czamara, N. Karbalai, C. von Toerne, M. Waldenberger, C. Gieger, P. Lichtner, M. Claussnitzer, R. Naumann, B. Müller-Myhsok, M. Torres, L. Garrett, Jan Rozman, M. Klingenspor, V. Gailus-Durner, H. Fuchs, M. Hrabě de Angelis, J. Beckers, S. M. Hölter, T. Meitinger, S. Hauck, H. Laumen, W. Wurst, F. Casares, J. Gomez-Skarmeta, J. Winkelmann (2014), Restless Legs Syndrome-associated intronic common variant in Meis1 alters enhancer function in the developing telencephalon. Genome Research, DOI: 10.1101/gr.166751.113