Researchers detect unwanted effects of important gene manipulation system
Researchers at the University of Virginia School of Medicine have devised a way to detect unintended side effects of manipulating genes using a revolutionary new system that is sweeping the scientific world by storm.
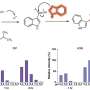
The gene targeting system, called CRISPR, allows editing of genetic information at specifically targeted sites in the genome. U.Va.'s new approach reveals the system has the potential to bind to unintended sites and cause gene mutations at some of these sites – mutations that could have serious consequences for research and efforts to develop medical treatments.
U.Va.'s new approach, however, also identifies ways to help prevent those potentially dangerous "off-target" effects, allowing scientists to improve their results with this important new gene-editing system.
Gene Mutations and How to Avoid Them
A primary goal of gene manipulation is to correct harmful mutations, so it is vital to avoid introducing mutations unintentionally, explained U.Va.'s Mazhar Adli, of the Department of Biochemistry and Molecular Genetics.
"We know that genetic mutations are hallmarks of disease. The whole aim is to change these apparent genetic mutations – to go and correct these mutations," he said. "We want to change this information only at the targeted space, at the targeted locus. If you change any other information, basically you are introducing mutations that you don't want. You are correcting one gene and potentially you might be introducing mutations in 10 other genes and maybe many other places in the genome."
CRISPR: The 'Holy Grail of Research'
Adli's new research sheds light on the potential off-target effects of the CRISPR/Cas9 gene editing system. The system has proved extremely popular because it allows scientists to manipulate specific sections of the genome of living mammals, making it a tool of tremendous importance for scientific research. It has been adopted quickly and widely, including for work in human cells, because it is comparatively simple and because many labs have the resources to use it.
"You can basically target any genomic region in living cells and change the genetic information, which has been the holy grail of research for the past several decades," Adli said. "To be able to go and change the genetic information in living cells was a dream, basically."
U.Va.'s new research helps explain the mechanism that underpins the CRISPR system – and why it is vulnerable to off-target gene mutations.
"We not only found where it binds in the genome, we also investigated why it goes to these regions in the genome," Adli said. "By analyzing specific sequences underlying these off-targets, we also found out the determinants – why it goes to the on-targets and also to these off-target regions – and our research shows that it goes there because of some sequence similarity to the original targeted regions.
"So our results will help improve the specificity of the system so that we can minimize the off-targets."
Major Mutation Implications
Adli's work also showed that the naturally occurring form of a key enzyme used in the CRISPR system introduces far more mutations than an altered form of that enzyme that is less commonly used. The former cuts both strands of DNA during gene editing, allowing mutations to occur, while the latter snips only one strand, allowing cells to repair the damage without introducing mutations.
"Unfortunately the wild-type form [the naturally occurring form] is much easier to deal with. Everyone in the field is using the wild type," he said. "Now with this paper, and with additional papers coming out, they will have to stop using the wild-type form of the enzyme. They have to use the [altered form] to overcome the off-targets. It is much superior and the off-targets are very dangerous."
Published Online
The findings have been published online by the journal Nature Biotechnology and will appear in a forthcoming print edition. The paper was written by U.Va.'s Cem Kuscu and Sevki Arslan, sharing credit as the lead authors; Ritambhara Singh; Jeremy Thorpe; and Adli.