Satellite Remote-Sensing Method Hatches New Cell-Analysis System
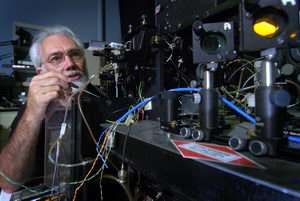
Using the same "multispectral analysis" concept that enables satellites to study Earth's surface, Purdue University researchers have developed a new system that quickly determines the composition of cells and tissue for medical diagnostics and scientific applications.
The method, which works by analyzing many separate colors in an object or surface, has been applied to the field of "flow cytometry," or analyzing cells that are contained in a liquid flowing past a laser beam, said J. Paul Robinson, a professor of cytomics in the School of Veterinary Medicine.
"Flow cytometry has been an important analytical tool for research and medicine for many years," said Robinson, who also is a professor in the Weldon School of Biomedical Engineering. "We have now modified this tool so that much more information can be gleaned in the same amount of time."
The new technique enables researchers to study 32 colors from a single cell flowing past a laser beam in a fraction of a second, promising to yield a wealth of data about cells for applications ranging from medicine to homeland security.
"Within five to 10 years, almost all of the instruments used in flow cytometry will have converted to this technology because it's smarter and better and cheaper," said Robinson, who is director of the Purdue University Cytometry Laboratories.
Recent research findings were detailed in a paper that was presented on Tuesday (May 23) during the 23rd International Congress of the International Society for Analytical Cytology.
In flow cytometry, a liquid treated with fluorescent dyes called "markers" flows past a laser beam at the rate of 10 meters per second.
"So, a single particle or cell is in front of the laser beam for about one ten-thousandth of a second as it flows by," Robinson said. "You couldn't blink your eye that fast. We are now able to make a full fluorescence profile of a single cell in that brief amount of time, meaning we can define vast numbers of properties of a cell."
Different markers automatically bind to specific cells, and the colored dyes glow when exposed to the laser beam. Analyzing a single particle or cell in 32 separate colors provides a "spectral signature" that enables researchers to diagnose disease or detect biological and chemical agents.
"Different diseases will be reflected by different proportions of certain types of cells," said Robinson, whose research is based at the Bindley Bioscience Center at Purdue's Discovery Park, the university's hub for interdisciplinary research.
The technique borrows complex computer algorithms developed decades ago by David Landgrebe, a Purdue emeritus professor of electrical and computer engineering.
"Forty years after David Landgrebe developed the fundamental tools, we are using the same technique in the realm of biology," Robinson said. "Using multispectral analysis with satellites is called remote sensing, and what we are doing with microscopes and cameras is called close-up sensing."
Satellites use multispectral analysis by viewing the same location on Earth with numerous filters to look at individual colors.
"You can determine the nature of a material by its spectral signature," Robinson said. "You can tell the difference between healthy wheat crops and wheat that has a fungus on it. You can tell the difference between land containing iron and land containing zinc because it has a different signature, and so on.
"Now, just transfer that to a microscope. We are doing exactly the same thing in tissue and cells."
Conventional flow cytometry technology, called polychromatic analysis, uses a series of mirrors to direct light into dozens of filters and detectors.
"One problem with this method is that the light becomes more and more dim as it is piped to the farthest detectors, so it becomes difficult to obtain high-quality data," Robinson said. "You also have a very complex mixing of signals, and you have to separate them using complicated mathematics."
Multispectral analysis uses a single mirror to direct light into a "holographic filter" that separates each of the 32 wavelengths and then into a detector that picks up each wavelength.
"Whereas conventional analysis requires very complicated optics, the multispectral cytometry has a relatively simple optical setup," Robinson said.
The new technique is especially useful if researchers have small amounts of samples to work with, such as the tiny amounts of blood available from premature babies.
"If you want to look at the contents of an adult's blood, it's no problem to take 10 milliliters of blood, but what do you do for a 4-pound neonatal infant?" Robinson said. "If you get one drop of blood, that's it, which means you can't do many of the tests that you would be able to do with an adult. You need to have a more sophisticated analytical tool that can get more information out of less material."
A more sophisticated analytical tool also is needed when analyzing rare types of cells and when trying to analyze many different types of cells within the same sample.
Flow cytometry enables scientists to analyze the blood to diagnose diseases and conditions ranging from AIDS to anemia. The tool also is used in homeland security to analyze the air for biological or chemical agents. Particles from the air are mixed with water, and then the water is analyzed.
The multispectral technique also could be used to analyze larger objects, such as the pattern of bacteria growing in a petri dish to detect food-borne contamination, and for other types of diagnostics.
"You could identify a certain kind of bacteria only a few hours after they started growing in the medium, which is much faster than you could with biochemical analysis," Robinson said. "This kind of fast diagnostics could be very helpful in not only identifying food pathogens but also to analyze the urine or other samples in medical microbiology."
The research paper being presented Tuesday was written by Robinson, research scientists Bartek Rajwa and Valery Patsekin, and researcher James Jones. Robinson is affiliated with the Purdue Cancer Center and the Purdue Oncological Sciences Center.
Source: Purdue University