The language of RNA decoded: Study reveals new function for pseudogenes and noncoding RNAs
The central dogma of molecular biology, as proposed in 1970 by Francis Crick and James Watson, holds that genetic information is transferred from DNA to functional proteins by way of messenger RNA (mRNA). This suggests that mRNA has but a single role, that being to encode for proteins.
Now, a cancer genetics team at Beth Israel Deaconess Medical Center (BIDMC) suggests there is much more to RNA than meets the eye.
In a study appearing in the June 24, 2010 issue of Nature, the authors describe a new regulatory role for RNA -- independent of their protein-coding function - that relies on their ability to communicate with one another. Of potentially even greater significance, because this new function also holds true for thousands of noncoding RNAs, the discovery dramatically increases the known pool of functional genetic information.
The new findings suggest that nature has crafted a clever tale of espionage such that thousands upon thousands of mRNAs and noncoding RNAs, together with a mysterious group of genetic relics known as pseudogenes, take part in undercover reconnaissance of cellular microRNAs, resulting in a new category of genetic elements which, when mutated, can have consequences for cancer and human disease at large.
"Because this new function does not depend on the blueprint that RNAs harbor in their protein-encoding nucleotide sequence, the discovery additionally holds true for the thousands of noncoding RNA molecules in the cell," explains senior author Pier Paolo Pandolfi, MD, PhD, Director of Research at the BIDMC Cancer Center and George C. Reisman Professor of Medicine at Harvard Medical School."This means that not only have we discovered a new language for mRNA, but we have also translated the previously unknown language of up to 17,000 pseudogenes and at least 10,000 long non-coding (lnc) RNAs. Consequently, we now know the function of an estimated 30,000 new entities, offering a novel dimension by which cellular and tumor biology can be regulated, and effectively doubling the size of the functional genome."
MicroRNAs are small RNA molecules that repress the expression levels of numerous genes by binding to mRNA, thereby preventing it from delivering its protein coding message. As a result, microRNAs are known to have a hand in human diseases, including cancer.
But, the Pandolfi team thought that RNA might be taking a different tactic. "Although it is conventional knowledge that microRNA can block mRNA function, we suspected that the roles of microRNA and mRNA were actually flipped," explains Pandolfi. "In other words, instead of microRNA binding to mRNA, we thought that RNAs were actually sequestering the microRNAs, thereby protecting mRNA and rendering the microRNAs ineffective on their other targets."
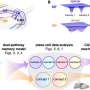
And, indeed this proved to be the case. The authors coined the term "competitive endogenous RNAs" or "ceRNAs" to describe this new RNA activity.
To further test their hypothesis, the scientists turned to pseudogenes, a group of "genetic relics" that do not encode for proteins. Since pseudogenes are more or less identical to their ancestral genes, the authors knew they would be the perfect "combatants" because they would both recognize and compete for the same group of microRNAs as their ancestral genes.
The scientists studied the interaction between the RNA encoding for the PTEN tumor suppressor gene and its closely related pseudogene, PTENP1. Through this new mechanism, they were able to demonstrate that PTENP1 also acts as a tumor suppressor. They also proved this to be true for the KRAS oncogene and its pseudogene KRAS1P; in this case the pseudogene was acting as a new oncogene.
"We now understand how these RNA units talk with one another," says Pandolfi. "Because all RNA molecules are competing to be heard, it had been difficult to tease out the messages amid the noise. But now that we know what to listen for, we can computationally predict which RNA can act as a ceRNA and what that ceRNA can do. And we can apply these findings to any RNA molecule.
"We have identified PTENP1 and KRAS1P as well as several thousand other uncharacterized RNA molecules as potential new factors contributing to human disease," he adds. "These findings help define a new fundamental biological dimension that we hope will allow for the rapid identification and functional characterizations of new disease-related genes including cancer, thereby improving diagnosis and effective therapy."