Unfolding amyloid secrets
Scientists from the University of Leeds have made a fundamental step in the search for therapies for amyloid-related diseases such as Alzheimer's, Parkinson's and diabetes mellitus. By pin-pointing the reaction that kick-starts the formation of amyloid fibres, scientists can now seek to further understand how these fibrils develop and cause disease.
Amyloid fibres, which are implicated in a wide range of diseases, form when proteins misfold and stick together in long, rope-like structures. Until now the nature of the first misfold, which then causes a chain reaction of misfolding by other proteins, was unknown.
Funded by the University of Leeds and The Wellcome Trust, the research published in Molecular Cell today is the culmination of four years of work led by Sheena Radford, Professor of Structural Molecular Biology and Deputy Director of the Astbury Centre for Structural Molecular Biology at the University of Leeds.
She explains: "We wanted to discover what happened to make a perfectly normal protein into one which was prone to aggregation because if we can stop the very first event, which causes a snowball effect, it provides us with new targets for future therapies."
The team first had to make a protein called beta-2 micro globulin, which when folded in a particular way is known to have a major role to play in the formation of amyloid fibres. These fibres particularly affect patients with kidney disease where they create deposits that can accumulate in the joints.
"Working kidneys get rid of beta-2 microglobulin," says Professor Radford. "But if you don't have properly functioning kidneys, you get a build up of the protein which can result in dialysis-related amyloidosis, which can be very painful."
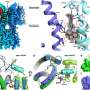
The researchers went on to solve the structure of this misfolded variant of beta-2 micro globulin - the first time its structure in its dangerous form has been directly shown. This allowed them to witness the properties that encourage other proteins to misfold and become amyloidegenic too.
Using nuclear magnetic resonance spectroscopy (NMR) to obtain high definition 3D images to view the structures, they found that only a small change or misfold in the protein made it unstable, causing it to become highly excitable and dynamic. This made it more likely to stick to other proteins, influencing their structure and starting off the snowball effect of aggregation.
"We saw that the variant protein bumped into others, stuck to them and changed their structure so that they too were amyloidegenic," says Professor Radford. "This is a huge step forward, not just for renal patients, but in our fundamental understanding of how amyloid fibres may form in other diseases as well. Many amyloid diseases are due to changes in protein structure and our next steps will be to see if similar changes are taking place with other protein types."