Researchers find mutation causing neurodegeneration
A Jackson Laboratory research team led by Professor and Howard Hughes Medical Investigator Susan Ackerman, Ph.D., has discovered a defect in the RNA splicing process in neurons that may contribute to neurological disease.
The researchers found that a mutation in just one of the many copies of a gene known as U2 snRNAs, which is involved in the intricate processing of protein-encoding RNAs, causes neurodegeneration.
Many so-called non-coding RNAs—those that don't directly encode proteins—are found in multiple copies in the genome, Ackerman says. "These copies are identical, or nearly identical, so conventional wisdom suggested they were redundant. For the first time, we show that a mutation in one copy can lead to disease."
The results, published in the journal Cell, suggest that disease-causing mutations may exist among other repetitive genes. "This opens up a whole new way of studying these RNAs," Ackerman notes, "including the types of disruptions in RNA processing that can lead to degeneration."
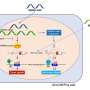
Neurons, like most other cells, build the workhorse proteins that carry out vital functions from the genetic "blueprint" encoded in DNA. In broad strokes, DNA gets copied by pre-messenger RNA (pre-mRNA), then pre-mRNA undergoes a splicing process before transporting the genetic code to the ribosome, where proteins are manufactured. But there's much more to it than that.
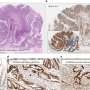
Specialized RNAs called U-snRNAs are essential to the splicing process. U-snRNAs are highly conserved, meaning that they are found all along the evolutionary pathway from simple organisms to humans. Ackerman showed that mutations in one form of snRNA, known as U2, lead to movement problems and early neuron death in mice.
U2 is a repetitive gene, meaning there are many copies of the same sequence. A mutation in just one copy led to the observed disorders by disrupting alternative splicing events, part of the splicing process that normally allows the creation of two or more protein forms from the same stretch of pre-mRNA.
The error leads to production of mRNAs containing regions known as introns that should have been removed. These abnormal mRNAs cause cell death, either through active toxicity or the production of dysfunctional proteins. Moreover, the researchers noted that the severity of the splicing abnormalities and cell death depend on the "dosage" level of the mutant gene.
Also, Ackerman and her lab noted that highest levels of the mutant U2 were found in the cerebellum, indicating that the expression of mammalian U2s, previously thought to be universal, may be different among various cell types.