This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
New class of cancer mutations discovered in so-called 'junk' DNA
Using artificial intelligence, Garvan Institute researchers have found potential cancer drivers hidden in so-called 'junk' regions of DNA, opening up possibilities for a new approach to diagnosis and treatment.
Non-coding DNA—the 98% of our genome that doesn't contain instructions for making proteins—could hold the key to a new approach for diagnosing and treating cancers, according to a new study from the Garvan Institute of Medical Research.
The findings, published in Nucleic Acids Research, reveal mutations in previously overlooked regions of the genome that may contribute to the formation and progression of at least 12 different cancers, including prostate, breast and colorectal.
The discovery could lead to early diagnosis and new treatments effective for many cancer types.
"Non-coding DNA was once dismissed as 'junk DNA' due to its apparent lack of function," says Dr. Amanda Khoury, Research Officer at Garvan and co-corresponding author of the study. "Our research has found mutations in these DNA regions that could open an entirely new, universal approach to cancer treatment."
Investigating DNA 'anchors' disrupted in cancer
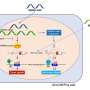
The researchers focused on mutations affecting binding sites for a protein called CTCF, which helps fold long strands of DNA into specific shapes. In their previous work, they found that these binding sites bring distant parts of the DNA close together, forming 3D structures that control which genes are turned on or off.
"We had already identified a subset of CTCF binding sites that are 'persistent'—that is they act like anchors in the genome, present across different cell types," says Dr. Khoury. "We hypothesized that if these anchors become faulty, it could disrupt the normal 3D organization of the genome and contribute to cancer."
To test this, the researchers developed a new sophisticated machine learning (AI) tool called CTCF-INSITE, which used genomic and epigenomic features to predict which CTCF sites are likely to be persistent anchors in a total of 12 cancer types.
They then assessed more than 3,000 tumor samples from patients diagnosed with the 12 cancer types, available from the International Genome Consortium database, and found the persistent anchors were rich with mutations.
"Using our machine learning tool, we identified persistent CTCF binding sites in 12 different cancer types," says Dr. Wenhan Chen, first author of the study. "Remarkably, we found that every cancer sample had at least one mutation in a persistent CTCF binding site."
"This research confirmed that persistent CTCF binding sites are 'mutational hotspots' in cancers. We think these mutations give cancer cells a survival advantage, allowing them to proliferate and spread," adds Dr. Khoury.
Towards a universal cancer treatment approach
The findings could have broad implications for understanding and treating many types of cancer.
"Most new cancer treatments have to be carefully targeted to specific mutations not always common among different tumor types, but because these CTCF anchors are mutated across multiple different cancer types, we're opening up the possibility of developing approaches that could be effective for multiple cancers," says Professor Susan Clark, Head of the Cancer Epigenetics Lab at Garvan and lead author of the study.
The researchers are now planning further large-scale experiments using CRISPR gene editing to investigate how these anchor mutations disrupt the 3D genome and potentially promote cancer growth.
"Now that we've discovered what we believe to be critical anchors of the genome and shown they are important to maintaining homeostasis of the genome architecture, it makes sense that these non-coding DNA mutations would disrupt this homeostasis in the cancer cell—a hypothesis we will test when we edit them out," says Professor Clark.
"Observing the downstream impact, we hope to identify key genes or gene pathways that are affected by the mutations, which could serve as markers for early cancer detection or targets for new treatments."
"Finding these clues that were hidden in a vast amount of data is a powerful example of how artificial intelligence is boosting medical research," she says. "This is a whole new frontier in the study of cancer, and we're excited to explore it further."
More information: Wenhan Chen et al, Machine learning enables pan-cancer identification of mutational hotspots at persistent CTCF binding sites, Nucleic Acids Research (2024). DOI: 10.1093/nar/gkae530