New way to learn about, potentially block traits in harmful pathogens
Researchers at Duke University Medical Center have developed a new way to identify the genes of harmful microbes, particularly those that have been difficult to study in the laboratory.
This new method uses chemicals to create mutant bacteria, followed by genomic sequencing to identify all mutations. By looking for common genes that were mutated in Chlamydia sharing a particular trait, the investigators were able to rapidly "zero in" on the genes responsible for that trait.
The approach is versatile and inexpensive enough that it could be applied to study a range of microorganisms, said Raphael Valdivia, Ph.D., an associate professor of molecular genetics and microbiology at Duke.
"We were able to learn about genes that allow Chlamydia to flourish in their hosts without the traditional, lengthy process of domesticating the pathogen to accept recombinant DNA," Valdivia said. "Our approach marries classical microbiology techniques with 21st century genome-sequencing technologies. If you encounter a new dangerous microorganism and want to determine what genes are important, I think this represents an effective way to learn all we can, as fast as we can."
One of the goals in studying microbial pathogens that harm humans and animals is to locate and disrupt the genes required for infection, Valdivia said.
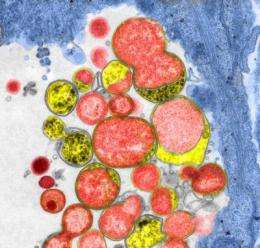
The microbe in this study, Chlamydia, is usually sexually transmitted, hides in human cells, and is a type of bacteria that must cause disease to be transmitted from one host to another. Chlamydia is the leading sexually transmitted infection and a risk factor for pelvic inflammatory disease and infertility.
Prior to this work, the function of many Chlamydia genes had to be inferred by their similarity to genes from other bacteria. "By isolating mutants that don't grow well inside cells and identifying the underlying mutations, we can learn a lot about how these genes contribute to disease," Valdivia said. "These are the activities we'd like to block."
"For us, this significantly accelerates the analysis of Chlamydia and importantly, should be applicable to many other microbes that have been difficult manipulate with recombinant DNA approaches," he said. Valdivia suggested that even microbes associated with our normal intestinal flora, which are notoriously difficult to manipulate, are now open to exploration so that we can learn how their genes influence human health, including dietary disorders and inflammatory bowel disease.
The work was published on Jan. 9 in the early edition of the Proceedings of the National Academy of Sciences.
Valdivia also said that the new technique could help to create Chlamydia vaccines that have a combination of mutations that affect the pathogen's virulence. "That way we can cripple some aspects of the bacterium's ability to thrive intact in a host, while still allowing the bacterium to replicate enough to prime the immune system against it."