New insight into why each human face is unique

The human face is as unique as a fingerprint, no one else looks exactly like you. But what is it that makes facial morphology so distinct? Certainly genetics play a major role as evident in the similarities between parents and their children, but what is it in our DNA that fine-tunes the genetics so that siblings – especially identical twins - resemble one another but look different from unrelated individuals? A new study by researchers at the U.S. Department of Energy's Lawrence Berkeley National Laboratory (Berkeley Lab) has now shown that gene enhancers - regulatory sequences of DNA that act to turn-on or amplify the expression of a specific gene – are major players in craniofacial development.
"Our results suggest it is likely there are thousands of enhancers in the human genome that are somehow involved in craniofacial development," says Axel Visel, a geneticist with Berkeley Lab's Genomics Division who led this study. "We don't know yet what all of these enhancers do, but we do know that they are out there and they are important for craniofacial development."
Visel is the corresponding author of a paper in the journal Science that describes this research. The paper is titled "Fine Tuning of Craniofacial Morphology by Distant-Acting Enhancers."
While some genetic defects responsible for craniofacial pathologies such as clefts of the lip or palate have been identified, the genetic drivers of normal craniofacial variation have been poorly understood. Previous work by Visel and his collaborators, in which they mapped gene enhancers in the heart, the brain and other organ systems demonstrated that gene enhancers can regulate their targets from across distances of hundreds of thousands of base pairs. To learn whether gene enhancers can also have the same long-distance impact on craniofacial development, Visel and a multinational team of collaborators studied transgenic mice.
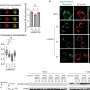
"We used a combination of epigenomic profiling, in vivo characterization of candidate enhancer sequences, and targeted deletion experiments to examine the role of distant-acting enhancers in the craniofacial development of our mice," says Catia Attanasio, the lead author on the Science paper. "This enabled us to identify complex regulatory landscapes, consisting of enhancers that drive spatially complex developmental expression patterns. Analysis of mouse lines in which individual craniofacial enhancers had been deleted revealed significant alterations of craniofacial shape, demonstrating the functional importance of enhancers in defining face and skull morphology."
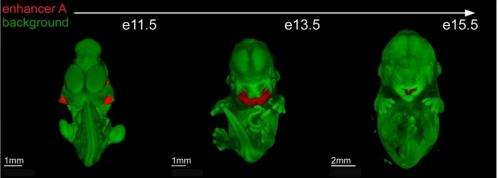
In all, Visel, Attanasio and their colleagues identified more than 4,000 candidate enhancer sequences predicted to be active in fine-tuning the expression of genes involved in craniofacial development, and created genome-wide maps of these enhancers by pin-pointing their location in the mouse genome. The researchers also characterized in detail the activity of some 200 of these gene enhancers and deleted three of them. A majority of the enhancer sequences identified and mapped are at least partially conserved between humans and mice, and many are located in human chromosomal regions associated with normal facial morphology or craniofacial birth defects.
"Knowing about the existence of these enhancers, which are inherited from parents to their children just like genes, knowing their exact location in the human genome, and knowing their general activity pattern in craniofacial development should facilitate a better understanding of the connection between genetics and human craniofacial morphology," Visel says. "Our results also offer an opportunity for human geneticists to look for mutations specifically in enhancers that may play a role in birth defects, which in turn may help to develop better diagnostic and therapeutic approaches."
Visel says he and his collaborators are now in the process of refining their genome-wide maps to gain additional information about the activity patterns of these enhancer sequences. They are also working with human geneticists to perform targeted searches for mutations of these enhancer sequences in human patients who have craniofacial birth defects.
More information: "Fine Tuning of Craniofacial Morphology by Distant-Acting Enhancers," by C. Attanasio; A.S et al. Science, 2013.