First large-scale PheWAS study using EMRs provides systematic method to discover new disease association
Vanderbilt University Medical Center researchers and co-authors from four other U.S. institutions from the Electronic Medical Records and Genomics (eMERGE) Network are repurposing genetic data and electronic medical records to perform the first large-scale phenome-wide association study (PheWAS), released today in Nature Biotechnology.
Traditional genetic studies start with one phenotype and look at one or many genotypes, PheWAS does the inverse by looking at many diseases for one genetic variant or genotype.
"This study broadly shows that we can take decades of off-the-shelf electronic medical record data, link them to DNA, and quickly validate known associations across hundreds of previous studies," said lead author Josh Denny, M.D., M.S., Vanderbilt Associate Professor of Biomedical Informatics and Medicine. "And, at the same time, we can discover many new associations.
"A third important finding is that our method does not select any particular disease - it is searches simultaneously for more than a thousand diseases that bring one to the doctor. By doing this, we were able to show some genes that are associated several diseases or traits, while others are not," he added.
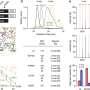
Researchers used genotype data from 13,835 individuals of European descent, exhibiting 1,358 diseases collectively. The team then ran PheWAS on 3,144 single-nucleotide polymorphisms (SNP's), checking each SNP's association with each of the 1,358 disease phenotypes.
As a result, study authors reported 63 previously unknown SNP-disease associations, the strongest of which related to skin diseases.
"The key result is that the method works," Denny said. "This is a robust test of PheWAS across all domains of disease, showing that you can see all types of phenotypes in the electronic medical record—cancers, diabetes, heart diseases, brain diseases, etc.—and replicate what's known about their associations with various SNPs."
An online PheWAS catalog spawned by the study may help investigators understand the influence of many common genetic variants on human conditions.
"If you think about the way genetic research has been done for the last 50 years or more, a lot of it was done through carefully planned clinical trials or observational cohorts," Denny said. "This certainly does not supplant those in any way but provides a cost efficient, systematic method to look at many different diseases over time in a way that you really can't do easily with an observational cohort."
Denny said PheWAS would be unworkable without the eMERGE Network, which has now expanded to nine sites with DNA samples from about 51,000 individuals linked to medical records. Vanderbilt is the coordinating center for eMERGE. The eMERGE Network is funded by the National Human Genome Research Institute.
"PheWAS opens up important avenues in understanding why certain diseases can present differently in different people, or how drugs might produce unpredicted effects in some patients," said senior author Dan Roden, M.D., assistant vice chancellor for Personalized Medicine, and principal investigator for the Vanderbilt eMERGE site.