Bioinformatics profiling identifies a new mammalian clock gene
Over the last few decades researchers have characterized a set of clock genes that drive daily rhythms of physiology and behavior in all types of species, from flies to humans. Over 15 mammalian clock proteins have been identified, but researchers surmise there are more. A team from the Perelman School of Medicine at the University of Pennsylvania wondered if big-data approaches could find them.
To accelerate clock-gene discovery, the investigators, led by John Hogenesch, PhD, professor of Pharmacology and first author Ron Anafi, MD, PhD, an instructor in the department of Medicine, used a computer-assisted approach to identify and rank candidate clock components. This approach found a new core clock gene, which the team named CHRONO. Their findings appear this week in PLOS Biology.
Hogenesch likens their approach to online profiling of movie suggestions for customers: "Think of Netflix. Based on your personalized movie profile, it predicts what movies you may want to watch in the future based on what you watched in the past." He thought the team could use this approach to identify new clock genes, given criteria already established from the "behavior" of known clock genes identified in the past two decades:
- Clock genes cause oscillations at the messenger RNA and protein level.
- Clock proteins physically interact with other clock proteins to form complexes that control daily rhythm inside cells.
- Disruption of clock genes in cell models cause changes in observable behavioral and metabolic traits on a 24-hour cycle.
- Clock genes are conserved across 600 million years of evolution from fruitflies to humans.
"We used a simple form of machine learning to integrate biologically relevant, genome-scale data and ranked genes based on their similarity to known clock proteins," explains Hogenesch. Using biological big data such as that found in the Circadian Expression Profile Data Base (CircaDB) to search for new clock genes, the Penn team evaluated the features of 20,000 human genes to isolate other genes that have the same clock-gene characteristics. "The hypothesis is that other genes that functionally resemble known clock genes are more likely to be clock genes themselves, just like movies that resemble your old favorites are more likely to become new favorites," says Anafi.
They found that several of the genes they identified physically interact with known clock proteins and modulate the daily rhythm of cells. One candidate, dubbed Gene Model 129, interacted with BMAL1, a well-known core clock component, and repressed the key driver of molecular rhythms, the BMAL1/CLOCK protein complex that guides the daily transcription of other proteins in a complicated system of genes that switch on and off over the course of the 24-hour day.
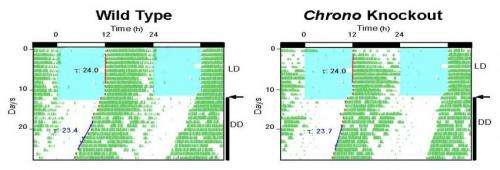
Given these results, the team renamed Gene Model 129, CHRONO, for computationally highlighted repressor of the network oscillator. The litmus test for identifying clock genes, however, is whether they regulate behavior: In mice in which CHRONO had been knocked out, Hogenesch found that the mice had a prolonged circadian period.
A companion study by colleagues at RIKEN in Japan and the University of Michigan, using a genome-wide analysis instead of a machine-learning approach, produced similar findings. Both studies link CHRONO to BMAL1. In the future, Anafi and Hogenesch will be investigating whether CHRONO regulates sleep, as most clock genes influence this behavior.