Powerful tool combs family genomes to find shared variations causing disease
Scientists at the University of Utah (U of U), the University of Texas MD Anderson Cancer Center in Houston and colleagues have developed a powerful tool called pVAAST that combines linkage analysis with case control association to help researchers and clinicians identify disease-causing mutations in families faster and more precisely than ever before.
In a study in Nature Biotechnology, the researchers describe cases in which pVAAST (the pedigree Variant Annotation, Analysis and Search Tool) identified mutations in two families with separate diseases and a de novo or new variation in a 12-year-old who was the only one in his family to suffer from a mysterious and life threatening intestinal problem.
"Linkage analysis and case control association traditionally have been used to find gene mutations," says Chad Huff, Ph.D., corresponding author on the study, assistant professor of epidemiology at the MD Anderson Cancer Center and former postdoctoral fellow in human genetics at the U of U. "Bringing those methods together provides a strong increase in the power to find gene variations that cause disease."
The advent of genome sequencing has allowed researchers to search for disease-causing mutations in the genomes of individual patients, larger groups of unrelated people or small and large families. The researchers in this study believe the most powerful way to identify these variants is by sequencing the genomes of families that experience unusually high occurrences of a particular illness. By identifying gene variations that family members share, it's possible to identify mutations in a gene that causes the disease, according to Mark Yandell, Ph.D., U of U professor of human genetics and a senior author on the paper.
"The issue with whole genome sequences has been that sequencing one person's genome to find a single disease-causing gene is difficult," Yandell says. "If you can sequence the whole family it gives a fuller picture of the sequence and variations potentially involved in disease."
Humans carry two healthy copies of each gene in the body. But mutations in a gene can cause disease or other health problems. These mutations occur randomly and rarely, but once they happen in a family member, they are often passed down to subsequent generations.
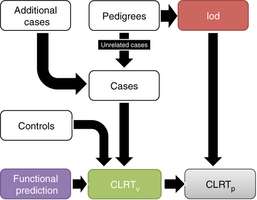
pVAAST was designed to search the sequenced genomes of families to find shared mutations and thus identify the gene with the highest probability of causing disease. Unlike other gene-finding tools, pVAAST accounts for people being related as it searches for gene variations that have the highest probabilities of causing disease. A big advantage of pVAAST, according to Huff and Yandell, is its ability to simultaneously search multiple families with the same disease to find mutations; this reduces the amount of time and effort to find a disease-causing variant. For example, if three families have the same disease, two might have different mutations damaging the same gene, while the third family might have a different damaged gene. "pVAAST has the power to determine the true disease-causing mutations across all those families in one analysis," Yandell says.
In related work, Yandell, Huff, and their colleagues vastly improved the results of individual and small family sequencing by developing another gene-finding tool, Phevor (Phenotype Driven Variant Ontological Re-ranking tool), which combines the probabilities of mutations being involved with a disease with databases of phenotypes and information on gene functions. In doing this Phevor and pVAAST in combination can identify disease genes with much greater precision than other tools.
Sequencing genomes of unrelated patients with the same disease also increases the ability to find gene variations, and a third software tool Yandell and colleagues developed, VAAST (Variant Annotation, Analysis and Search Tool), has greatly advanced the speed and precision of doing that.
If VAAST or pVAAST can't identify the mutation most likely to cause a disease, Phevor can take the results from those tools and combine them with a description of the patients' disease called a 'phenotype' to find the most likely causative gene.
"We hope that in developing pVAAST, we and other researchers can more rapidly identify genetic variations influencing disease risk by increasing the statistical power of familial genome sequencing," Huff says.
More information: A unified test of linkage analysis and rare-variant association for analysis of pedigree sequence data, Nature Biotechnology (2014) DOI: 10.1038/nbt.2895