Scientists reverse bacterial resistance to antibiotics
The rise of antibiotic-resistant bacteria is a growing problem in the United States and the world. New findings by researchers in evolutionary biology and mathematics could help doctors better address the problem in a clinical setting.
Biologist Miriam Barlow of the University of California, Merced, and mathematician Kristina Crona of American University tested and found a way to return bacteria to a pre-resistant state. In research published in the open-access journal PLOS ONE, they show how to rewind the evolution of bacteria and verify treatment options for a family of 15 antibiotics used to fight common infections, including penicillin.
Their work could have major implications for doctors attempting to keep patient infections at bay using "antibiotic cycling," in which a handful of different antibiotics are used on a rotating basis.
"Doctors don't take an ordered approach when they rotate antibiotics," Barlow said. "The doctors would benefit from a system of rotation that is proven. Our goal was to find a precise, ordered schedule of antibiotics that doctors could rely on and know that in the end, resistance will be reversed, and an antibiotic will work."
Dangers of Antibiotic Resistance
When bacteria grow powerful enough that antibiotics no longer work, it can be a matter of life and death. Recently, at the Ronald Reagan UCLA Medical Center, two people died and seven were injured when a medical scope used in patient procedures harbored drug-resistant bacteria. In the U.S. annually, more than 2 million people get infections that are resistant to antibiotics and at least 23,000 people die as a result, according to the Centers for Disease Control and Prevention.
Resistance to antibiotics is a natural part of the evolution of bacteria, and unavoidable given the many types of bacteria and the susceptibility of the human host. To compensate for bacterial evolution, a doctor fighting infections in an intensive care unit may reduce, rotate or discontinue different antibiotics to get them to be effective in the short term.
The researchers—from UC Merced, AU and UC Berkeley—have been leading the way to uncover how to reverse resistance in the drug environment. They've done so by combining lab work with mathematics and computer technology.
"We have learned so much about the human genome as well as the sequencing of bacteria," Crona said "Scientists now have lots and lots of data, but they need to make sense of it. Mathematics helps one to draw interpretations, find patterns and give insight into medical applications."
Challenging Work Yields Important Results
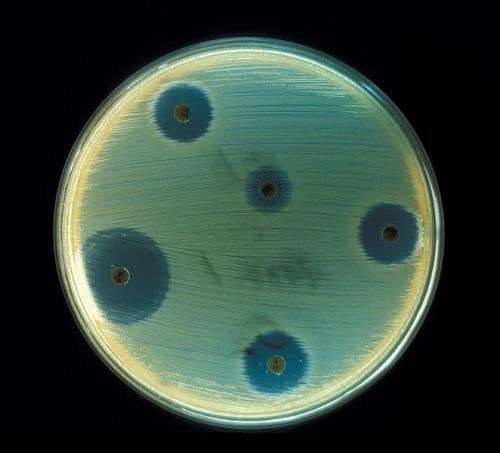
After creating bacteria in a lab, the researchers exposed them to 15 different antibiotics and measured their growth rates. From there, they computed the probability of mutations to return the bacteria to its harmless state using the aptly named "Time Machine" software.
Managing resistance in any drug environment is extremely difficult, because bacteria evolve so quickly, becoming highly resistant after many mutations. To find optimal cycling strategies, the researchers tested up to six drugs in rotation at a time and found optimal plans for reversing the evolution of drug-resistant bacteria.
"This shows antibiotics cycling works. As a medical application, physicians can take a more strategic approach," Crona said. "Uncovering optimal plans in antibiotics cycling presents a mathematical challenge. Mathematicians will need to create algorithms that can deliver optimal plans for a greater amount of antibiotics and bacteria."
The researchers hope to next test the treatment paths in a clinical setting, working with doctors to rotate antibiotics to maximize their efficacy.
"This work shows that there is still hope for antibiotics if we use them intelligently," Barlow said. "More research in this area and more research funding would make it possible to explore the options more comprehensively."
More information: PLOS ONE, dx.plos.org/10.1371/journal.pone.0122283