June 2, 2015 feature
From gene to phene: Scientists demonstrate genetic control of phenotypic variability
(Medical Xpress)—One of the most challenging problems in biology is the extreme difficulty in predicting phenotype from genotype – and the questions it creates: If we could rear genetically identical individuals from a variety of genetic backgrounds and rear them in the same environment, how much phenotypic variation between individuals of the same genotype would we see? Would different genetic backgrounds differ in their degree of variability? What would account for these differences? Recently, scientists at Harvard University, Cambridge and Cornell University, Ithaca used Drosophila (fruit flies) inbred lines to address these questions focusing on variability in locomotor handedness. They demonstrated that different genotypes vary significantly in phenotype variability; that this phenotypic variability itself, as a trait, can be heritable; and that genomic locations affecting variability can be mapped. Moreover, taken together with a companion study1, the papers demonstrate a rare example of linkage between genetic variation for a complex behavioral trait and a neural center of behavioral control.
Dr. Julien F. Ayroles discussed the paper that he, Dr. Benjamin de Bivort, and their colleagues published in Proceedings of the National Academy of Sciences. "Our work was motivated by the realization that nearly all genetic projects using genome wide association studies to map genes associated with various traits or diseases focus on detecting average differences between alternative alleles – that is, DNA polymorphism," Ayroles tells Medical Xpress. (DNA, or genetic, polymorphism refers to the occurrence in the same population of two or more alleles – variant forms of a gene – at the same location.)
If one takes a large population of individuals, Ayroles illustrates, aggregates all individuals with a thymine nucleotide at a given base pair and, separately, all individuals with a guanine nucleotide, and then compares, for example, the average height of the thymine and guanine populations, a difference in height between the two groups suggests that this DNA segment is associated with height. "However, we now know that sequence variation can also control the variance of a trait, in addition to the mean." In this example, thymine may be associated with low variance and guanine with high variance. "One of the major goals of genomics is to explain how sequence variation between individuals is associated with the dramatic phenotypic variation we observe between individuals, and we'd ultimately like to be able to predict phenotype from genotype. Unfortunately, at this point we're falling short of this goal: Genetic control of variance appears to be far more important than preciously appreciated and we know virtually nothing about how it works. Our study is first in that direction."
In discussing the use of Drosophila (fruit fly) inbred lines in their research, Ayroles points out that the idea that genomes not only encode trait means but also their degree of variability is an old observation. "Animal breeders, who commonly work with inbred stocks, have long noticed that different individuals of the same genotype can differ dramatically, and that some genotypes are more variable than others. That said, studying the biology of this phenomenon has been very difficult – it requires model systems that can be studied in the lab and genetically manipulated. Drosophila is ideal to work on this problem: We already have large panels of inbred lines, we can rear very large numbers of individuals for a large collection of genotypes to estimate variance, we have high throughput assays, and genomic sequences are available for a large number of Drosophila lines, allowing gene mapping."
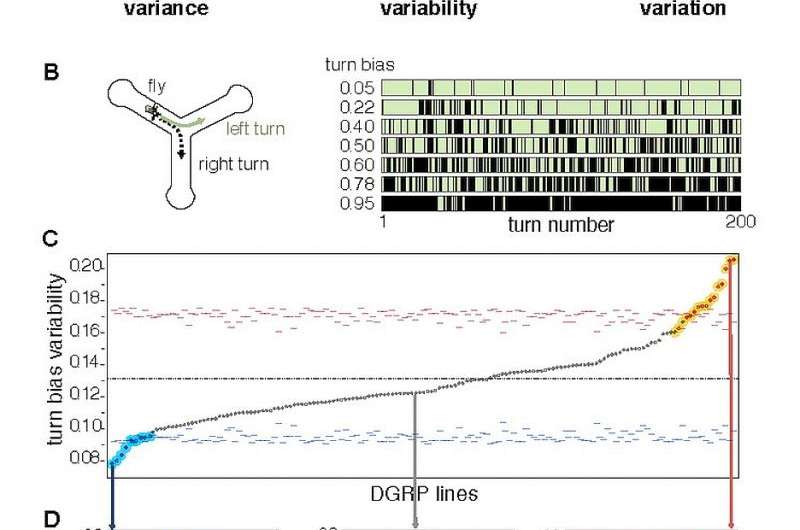
In essence, the study conducted by Ayroles and his colleagues was designed to address the question of how much phenotypic variation between individuals of the same genotype they would see if they could "clone" individuals from a variety of genetic backgrounds and rear them in the same environment – if such variation was found, what would account for these differences, "This is more or less what we did," Ayroles tells Medical Xpress. "We reared about 100 Drosophila individuals for each of almost 200 inbred lines – that is, genotypes – and for each individual across lines we measured the simple behavioral trait of locomotor handedness in a Y-shaped maze, which is useful because left/right decisions can be measured without experimental error." Figure 1 shows that on average, all the lines are even handed, meaning each population of 100 flies (per line), on average goes left/right with equal probability (50/50) – which Ayroles notes is true for all the genotypes they assayed. "However, what we also noticed is that the distribution of turning bias between lines differed dramatically, meaning that some genotypes had low levels of variability around the 50/50 mean while other lines had a high degree of variability. As a result, in high-variability genotypes, we were more likely to encounters individuals with an extreme turning bias – for example, with flies going to the left 80% of the time over several hundred turning decisions."
Ayroles adds that there's yet another aspect to genetic variability. "The fact that the average handedness is 50/50 for all the lines means that handedness itself is not under genetic control: If you were to take a male and female fly with a strong left turning bias and crossed them to produce offspring, their progeny will not have a left turning bias – they will revert to the 50/50 mean. However, as a trait, variability itself is heritable, so that if you cross a male and female, each from a high variability genotype, their progeny will have high variability and the same applies to low variability." This means that the property that is inherited is not the turning bias itself, but rather its variability. "We now have an experimental system allowing us to study the genetics of variance control without confounding the signal with genetic control of the mean. As far we know this is a first."
Having established that variability, as trait, was heritable, the scientists proceeded to map the genes associated with differences in variability. "Since the genomes of all the Drosophila lines we used have previously been sequenced, we performed a genome-wide association study" – an examination of many common genetic variants in different individuals to see if any variant is associated with a trait. "We looked for correlations between a given sequence variant known as a single-nucleotide polymorphism, or SNP, and our trait of interest – in this case, variability. We found a few genes associated with variability without affecting the mean, but one in particular was very interesting – a gene called Ten-a, which affects the fly's brain central complex, which is involved in locomotor decision-making."
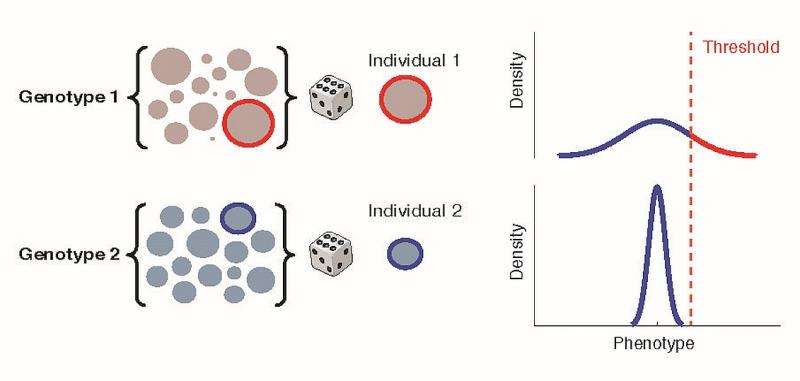
One of the key goals of quantitative genetics is to determine and describe the fraction of phenotypic variation between individuals attributable to genetic factors – in short, to measure heritability and explain heritability at the genetic level. "The traditional approach used to study the genetic basis for variation in any complex trait – in humans for example – is currently to run a genome-wide association study focused on the mean," Ayroles explains. "The design is simple: sample a number of individuals, measure a phenotype of interest for each of them, assay genotypes, and determine whether or not there is an association between the two. This Drosophila experiment, however, really shows the benefit of examining phenotypic variability among same-genotype individuals and the ability to study diversity usually hidden in population averages. Imagine running a genome wide association study in the flies the same way it would be run for a human study, where instead of measuring 100 flies of the same genotype across a large number of genotypes, we sampled only a single fly per genotype for a large collection of genotypes. While variation in handedness would still be observed in the collection, this genome-wide association study would not be able to identify any genes associated with variation in handedness - and we now know why: All the variation in handedness between individual flies in this one-fly-per-genotype sample is attributable to differences in variability between lines rather than in the mean. The differences between individuals are therefore explained by their degree of variability, not a deterministic mean attribute encoded in their DNA. Hence, if we did not focus on variance we would have missed this link between genotype and phenotype."
In order to make statistically clear distinctions between genetic and environmental effects on variability, the researchers took two steps, Ayroles recounts. "First, experimentally, we made sure that the environment is as constant as possible and – more importantly – properly randomized." This means that flies from each genotype are measured over several batches, and the scientists ensure that the effect they measure is not a function of that batch. "Secondly, statistically, this is not any different than any calculation of heritability, which is defined as the proportion of the total phenotypic variation in a population that can be attributed to genetic variation. Given this design, we can investigate the consistency of the variability within a genotype compared to the variation in variability between genotypes."
Moreover, Ayroles tells Medical Xpress that some unidentified effect is causing variation between individuals within a line, given that they have the same genotype. "These differences are most likely caused by subtle environmental perturbations called microenvironmental factors – for example, variations in molecular gradients during development – as opposed to macroenvironmental perturbations such as diet or temperature. The differences between high- and low-variability genotypes are most likely explained by variation in the differential ability of some genotype to buffer these microenvironmental perturbations."
When asked if differences in phenotypic variance between genotypes has implications for evolutionary biology, Ayroles stresses that it is again about predictability. "If selection optimizes a genotype to a given environment but the phenotypic variance of that genotype is high, by chance alone this genotype may produce 'misfits.' This of course could be bad in a stable environment, as it would lower the fitness of individuals carrying high variance alleles – but if the environment is highly variable and unpredictable, a genotype which by chance produces a wider array of phenotypes may be more successful than one narrowly adapted to a given environment. The literature refers to this as bet-hedging." (Evolutionary bet-hedging is a response to environmental variability in which organisms adapt their physiology or behavior to maximize survival probability.)
Ayroles points out that the current study has important implications for medical genetics. "As mentioned, one of the most vexing problems in this genomic area is our very poor performance at predicting phenotype from genotype. The leading paradigm is that we live in an additive/linear world, where if we knew all the genes and alleles affecting a trait we'd be able to make accurate predictions – but we are not able to do so. Our study shows that variance control may play a role in this poor performance."
There also appears to be a relationship between intragenotypic variability in personality (defined broadly for a wide range of species in terms of animal behavior) and neurobiology. A companion paper1 by Dr. Sean Buchanan published in the same issue of PNAS mapped a set of neurons a sensory integration and motor coordination center within the central complex – a part of the Drosophila brain that regulates the magnitude of left-right turn bias and therefore the magnitude of intragenotypic variability – using the same assay and traits as did Ayroles and his colleagues. The paper published by Ayroles and his colleagues states that taken together, the two studies "constitute a rare example linking natural genetic variation for a complex behavioral trait, to mutants implicating a brain region, to a specific subcircuit within this region. Thus, we can begin to paint the path from genetic variation to behavioral individuality."
As to their ongoing research, the scientists are now studying additional behavioral traits to better understand how much variation between individuals can be attributed to genetic control of variability, and are extending this analysis to other model systems, including mice and humans. "We're trying to determine the contribution of variability to our inability to make reliable genotype-to-phenotype predictions," Ayroles emphasizes.
While the genetics of variance control is difficult to study, and inbred lines are one of the most powerful tools available, Ayroles is also working on achieving understanding of the causes and consequences of genetic control of variance beyond behavior: Using a large cohort of twins, Ayroles says they can ask whether some DNA polymorphisms are associated with the probability of concordance or discordance between twin pairs (for example, SNP A twins are more likely to be similar to each other for a given trait, and SNP T twins are likely to have variable trait values). "We're also investigating this problem in humans using large families based on pedigrees."
Regarding areas of research that might benefit from their study, Ayroles says that "there's no doubt that this work speaks to our understanding of determinism in biology. He also states that one of the most important areas where this work may matter is in pharmacology. "One the typical steps in the drug development experimental pipeline is exposing a population of inbred mice to the drug – and one can imagine how different the conclusion would be if one is working with a high variability inbred line compared to a low variability line. While the means may or may not be the same, the variance really matters – especially in cases where the tail of the distribution is an 'unhealthy' zone." Ayroles cautions that while there are many examples of this in the literature, very few people are paying attention.
In a wider sense, Ayroles concludes that in systems from engineering to ecosystems, maintaining parameters within a certain range is very critical – and for that reason most systems must have some buffering mechanisms to control variance as an optimal mean.
More information: Behavioral idiosyncrasy reveals genetic control of phenotypic variability, Proceedings of the National Academy of Sciences (2015) 112(21):6706-6711, doi:10.1073/pnas.1503830112
Related:
1Neuronal control of locomotor handedness in Drosophila, Proceedings of the National Academy of Sciences (2015) 112(21):6700-6705, doi:10.1073/pnas.1500804112
© 2015 Medical Xpress