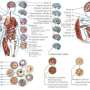
Scientists build genome-based 'reference library' for blood diseases
A new paper on how genome data (information regarding the genetic material of an organism such as DNA) can be made open-access – while at the same time ensuring that appropriate levels of privacy are maintained – has been published. The paper contains guidelines designed to both reduce ambiguity in the interpretation of open-access data and achieve controlled access.
The paper was written by researchers from several major research projects including the EU-funded BLUEPRINT project. This major five year initiative, which was launched in October 2011, will receive nearly EUR 30 million in EU funding. The project could transform European healthcare delivery by furthering our understanding of how genes are activated or repressed in both healthy and diseased human cells.
It is hoped that the results of the BLUEPRINT project will lead to better targeted diagnostics and new treatments for specific blood-related diseases in individual patients, an approach known as 'personalised medicine'. In addition, the involvement of innovative companies will energise epigenomic research in the European private sector, through the development of new smart technologies.
Disease, damage and a host of environmental factors including stress and diet can cause changes in DNA in the nucleus of a cell. These so called epigenetic alterations are not actual modifications in the genetic code, but involve the addition of chemical groups to proteins involved in DNA organisation. The placement of these has been associated with diseases such as cancer and diabetes. It is this process that BLUEPRINT – which focuses on blood cells –hopes to better understand.
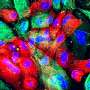
The project began by collecting data on how genes are activated or repressed. The aim here is to generate 100 reference epigenomes (records of chemical changes to DNA) to help the team identify markers or indicators of changes that might herald stages of disease. Data on a number of healthy epigenomes from different blood cell types have already been published, leading to the identification of several new epigenetic modulators involved in tumour development.
In order to cope with the enormous amounts of data, a common data coordination and analysis strategy has been established, along with frameworks for controlled data access. This infrastructure enables BLUEPRINT to make epigenomic maps immediately available to the scientific community. Better diagnosis tools are also being developed, which should reduce the need for huge quantities of cells.
Finally, the project's goals fit very much into the overall objective of the International Human Epigenome Consortium (IHEC), a global consortium that aims to encourage free access to high-resolution reference human epigenome maps for the research community. BLUEPRINT will make a valuable contribution here by generating reference epigenomes through the use of state-of-the-art technologies, all in accordance with quality standards set by IHEC.
The BLUEPRINT project is scheduled for completion in September 2016.
More information: For further information, please visit BLUEPRINT: www.blueprint-epigenome.eu/