Researchers generate a reference map of the human epigenome

The sequencing of the human genome laid the foundation for the study of genetic variation and its links to a wide range of diseases. But the genome itself is only part of the story, as genes can be switched on and off by a range of chemical modifications, known as "epigenetic marks."
Now, a decade after the human genome was sequenced, the National Institutes of Health's Roadmap Epigenomics Consortium has created a similar map of the human epigenome.
Manolis Kellis, a professor of computer science and a member of MIT's Computer Science and Artificial Intelligence Laboratory and of the Broad Institute, led the effort to integrate and analyze the datasets produced by the project, which constitute the most comprehensive view of the human epigenome to date.
In a paper published today in the journal Nature, Kellis and his colleagues report 111 reference human epigenomes and study their regulatory circuitry, in a bid to understand their role in human traits and diseases.
"The consortium set out to systematically characterize the human epigenomic landscape, across diverse tissues and cell types," Kellis says. "Given the enormity of the task, that meant bringing together multiple mapping centers and profiling a wide range of cell and tissue samples, to capture the diversity of the human epigenome."
150 billion genomic sequences
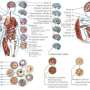
The researchers generated 2,805 genome-wide datasets, encompassing a total of 150 billion sequencing reads, corresponding to 3,174-fold coverage of the human genome. These captured modifications of both the DNA itself, and of the histone proteins around which DNA is wrapped to form a structure known as chromatin.
Kellis and his team then developed and applied machine-learning algorithms that could translate these datasets into a reference map in each of the 111 cell types and tissues. The algorithms distinguished different classes of epigenomic modifications and used them to annotate the genomic regions active in each sample, and in particular regulatory elements that control where and when different genes are expressed.
"Different combinations of epigenetic marks characterize different regions of the genome, reflecting the specific functions that they play in each cell," Kellis says. "By studying these combinations systematically, we can learn the language of the epigenome, and what it is telling us about both the activity and the function of each genomic region in each of the cell types."
The researchers distinguished 15 different epigenomic signatures, or chromatin states, reflecting active, repressed, poised, transcribed, and inactive regions of the genome in each cell type. About 5 percent of each reference epigenome showed signatures associated with a regulatory function.
"Chromatin states allowed us to summarize the complexity of diverse epigenomic marks into a small number of common patterns," Kellis says. "We could then interpret the biological functions of these patterns."
Epigenomic dynamics
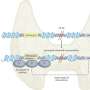
The researchers then studied how these chromatin states varied across different types of cells and tissues. This allowed them to group cell types with similar regulatory circuitry. They also grouped together regulatory regions that are active in the same types of cells. In this way they could begin to reveal the building blocks of regulatory circuits.
"Unlike the genome, which is mostly unchanged across cell types, the epigenome is extremely dynamic, reflecting the specialization of each cell type, such as neurons, heart, muscle, liver, skin, blood, or immune cells," Kellis says. "By studying which regions turn on and off in the same cell types, we can gain insights into gene regulation."
The researchers grouped 2 million predicted regulatory regions into 200 sets, or modules, which appeared to be acting in a coordinated manner across different types of cells. They found that 100 of these modules contained common sequence patterns, known as regulatory motifs, which may be responsible for their ability to work together in this way.
"Exploiting the predicted regulators and their motifs can help dissect the circuitry of different tissues and cells," Kellis says.
The researchers also compared these epigenomic signatures with groups of genetic variants that are associated with different human traits and diseases. This allowed them to produce a map of the tissue and cell types that are most relevant to each trait or disease.
"We found that genetic variants are found in regulatory regions known as enhancers, which are activated only in certain types of cell and tissue," Kellis says. "This suggests that many genetic variants affect the regulatory circuitry of the cell, possibly disrupting gene functions by altering tissue-specific gene expression levels."
Tissue-specific enhancers affect 58 traits
The researchers found significant tissue-specific enhancer signatures for genetic variants associated with 58 different traits. These included height, in embryonic stem cells; multiple sclerosis, in immune cells; attention deficit disorder, in brain tissues; blood pressure, in heart tissues; fasting glucose, in pancreatic islets; cholesterol, in liver tissue; and Alzheimer's disease, in CD14 monocytes.
"This unbiased view allows researchers to focus on relevant cells and tissues that may have been otherwise overlooked when studying a particular disease," Kellis says. "The regulatory circuitry of a diverse range of cells can contribute to diseases that manifest in seemingly unexpected organs."
Using these circuits to understand the molecular basis of human disorders will take many years and the effort of many labs, Kellis says. "Our results provide an invaluable map, and a rich set of hypotheses, which can help guide these studies."
Wolf Reik, head of the epigenetics research program at the Babraham Institute in the U.K., who was not involved in the research, says the project is an exciting resource for the biomedical community worldwide.
.
"Important epigenetic marks were mapped systematically in many human cell types and tissues," Reik says. "Integrative analysis of these epigenomes provides a global map toward understanding fundamental developmental and disease processes in humans."
More information:
DOI: 10.1038/nature14248
DOI: 10.1038/nature14252
DOI: 10.1038/nbt.3157
The research will be freely available to all, and the papers from Nature, along with papers from Nature Communications, Nature Biotechnology, Nature Methods, Nature Neuroscience, Nature Immunology and Nature Protocols, can be explored on the Epigenome Roadmap Site at www.nature.com/epigenomeroadmap.
This story is republished courtesy of MIT News (web.mit.edu/newsoffice/), a popular site that covers news about MIT research, innovation and teaching.