Researchers develop hybrid computational strategy for scalable whole genome data analysis
Human genome sequencing costs have dropped precipitously over the last few years, however the analytical ability to meet the growing demand for making sense of large data sets remains as a bottleneck. With the introduction of 'leaner and meaner' sequencers several years ago, there has been an exponentially increasing need to expand the number of human genomes and data volume for biomedical studies, both academically and commercially. Computational solutions, both software and hardware, are severely needed.
In a study published in BMC Bioinformatics, researchers from Baylor College of Medicine's Human Genome Sequencing Center, along with Oak Ridge National Laboratory, DNAnexus and the Human Genetics Center at the University of Texas Health Science Center, have developed a novel hybrid computational strategy to address this challenge when processing large data sets.
This new strategy has proven successful in analyzing an unprecedented set of 5,000 samples, which constitute a critical part for the international consortia efforts called CHARGE (The Cohorts for Heart and Aging Research in Genomic Epidemiology), aiming to identify genetic culprits for a number of common chronic diseases.
The computational solutions described in the paper ensure a timely delivery, in six weeks, of high quality genetic variant results to hundreds of scientists worldwide, who anxiously wait to work on the variant datasets, which might lead to breakthroughs and identifying variants and genes accountable for higher risks of heart diseases and diabetes.
Quality assurance
The study addressed three major challenges in large scale sequencing projects; maintaining high quality when sequencing a highly heterogeneous set of many thousands of samples, pulling and analyzing results in a timely manner and creating a scalable process for increasingly large data sets.
"There was previously no infrastructure for this large of a set, at 5,000 samples," said Dr. Eric Boerwinkle, associate director of Baylor's Human Genome Sequencing Center and dean of UT Health School of Public Health. "To address this, we employed a combination of platforms to perform large-scale variant calling, while maintaining high quality data."
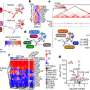
The computing framework utilizes cloud Amazon Web Services (AWS) for joint calling of single nucleotide variants (SNV), and supercomputers and local high performance computing infrastructures to carry out the imputation and phasing. These platforms were located at Baylor, Oak Ridge National Laboratory and Rice University, respectively.
This novel combination of platforms can be employed to scale up to 10,000 samples while producing SNV call sets with high sensitivity and specificity. In this particular study, the team was able to execute the joint calling, imputation and phasing of more than 5,300 whole genome samples in six weeks, using four callers, including SNPTools, GATK-Haplotype Caller, GATK-UnifiedGenotyper and GotCloud. The operation as a whole used 5.2 million core hours, and transferred 6 terabytes of data across the platforms.
"This is an excellent example of two scientific communities coming together to address challenging science problems. We are happy to have played a part in conducting the analysis of such unprecedented scale," said Dr. Manjunath Gorentla Venkata, co-author and computer scientist at the Department of Energy's Oak Ridge National Laboratory. "While researchers from BCM discussed the problem, we did not have a ready-made solution. After multiple discussions, we were convinced that mapping pipeline components based on system architecture strengths and tailoring parameters to the architecture would provide quality analysis with a relatively short turnaround time. "
"The Oak Ridge Leadership Computing Facility (OLCF), a DOE Office of Science User Facility, hosts the most capable supercomputer and data infrastructure for science in the United States", said Jack Wells, director of science of the OLCF. "Our facilities are well suited to support pioneering user projects, for example whole genome data analysis, and more breakthrough science across a wide spectrum of science domains."
Forward thinking
"The demand for and the sheer size of sequencing is advancing more quickly than the downstream analytical technologies can adapt." said Dr. Zhuoyi Huang, the leading author and a postdoctoral fellow with Baylor's Human Genome Sequencing Center.
"We have created a strategy that is highly scalable for increasingly larger samples, and have developed an understanding of best practices for the process, which can be replicated by other research institutions," said Dr. Navin Rustagi, the other leading author on the paper, also a postdoctoral fellow with the Human Genome Sequencing Center at Baylor.
This strategy can accomplish the ensemble joint calling of SNVs in a scalable, cost effective and timely manner, without compromising the quality of variants, by combining heterogeneous computing platforms.
"It has been a tremendous experience to work with a Cloud provider such as DNAnexus, as well as Oak Ridge National Laboratory and Rice University, applying their cloud platforms and supercomputer facilities to address various computational challenges across the entire schema," said Dr. Fuli Yu, assistant professor at Baylor's Human Genome Sequencing Center and senior author on the paper. "The excitement in this research is not only the scale, but also the interdisciplinary nature in the various levels of this operation."
More information: Zhuoyi Huang et al. A hybrid computational strategy to address WGS variant analysis in >5000 samples, BMC Bioinformatics (2016). DOI: 10.1186/s12859-016-1211-6