Study outlines framework for identifying disease risk in genome sequence

Imagine a day when you visit the doctor's office for your annual physical. Your physician orders routine tests - cholesterol, glucose and blood count - but they also order a sequence of your genome, all 3 billion letters of it. Routine genomic testing is not far away, according to researchers at The University of Texas Health Science Center at Houston (UTHealth).
"Whole genome sequencing will become an integral part of routine medicine in the near future," said Eric Boerwinkle, Ph.D., dean, M. David Low Chair in Public Health and Kozmetsky Family Chair in Human Genetics at UTHealth School of Public Health.
To help physicians, patients and scientists process this incredible amount of data, Boerwinkle and his team of genetic researchers have developed a framework to understand how whole genome sequence data can be analyzed to identify genetic variations that raise or lower risk of disease. A paper on the framework was published today in the American Journal of Human Genetics.
"The important aspect of this work is that it provides practical steps for scientists and physicians to help analyze whole genome sequences to identify differences that may be increasing disease risk or protecting certain individuals from diseases such as diabetes, cancer and heart disease," said Alanna Morrison, Ph.D., professor and chair of the Department of Epidemiology, Human Genetics and Environmental Sciences at the School of Public Health. Morrison was first author of the study.
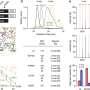
In the paper, researchers sequenced the genomes of more than 3,000 people from the Atherosclerosis Risk in Communities (ARIC) study and analyzed multiple traits related to heart and blood disease.
The framework they developed allowed them to examine different functional parts of the genome, including the genes that code for proteins and the portions of the genome that are used to control the expression of genes. "It allowed us to pull apart the different functional components of genome, which we couldn't access before comprehensively," said Morrison.
Through the framework, researchers were able to identify genes for blood lipid levels, white blood cell count and a molecule, troponin, which helps diagnose heart attacks.