TB bacteria evolve at alarming rate

Researchers from the Vavilov Institute of General Genetics of the Russian Academy of Sciences (VIGG) and the Moscow Institute of Physics and Technology (MIPT) have established a catalog of mutations in 319 virulence genes of mycobacteria that cause tuberculosis. These genes encode proteins that suppress human immune response. Further analysis has identified a set of three mutations which may enable mycobacteria to develop rapidly in an immunocompromised environment. The emerging strains of TB pathogens require new treatment approaches, including the development of new genetically engineered vaccines that take into account both the immune status of a patient and the specific virulence features of a pathogen. The article was published in Genome Biology and Evolution.
Lost battle
According to the World Health Organization, TB remains one of the most dangerous human infectious diseases, causing over 1.8 million deaths annually. TB is caused by a bacterium known as Mycobacterium tuberculosis or Koch's bacillus. It is clear that HIV-positive individuals and patients with other immunodeficiency conditions are particularly at risk. Additionally, more than 20 percent of TB cases are connected with smoking. TB is no longer a social disease, affecting members of all social strata, a result of the stresses of modern life.
M. tuberculosis has become increasingly resistant to both the environmental factors and antibiotics that used to guarantee effective treatment. At the same time, the symptoms of TB have become less noticeable. The bacterium can remain in the host body for decades, infecting other people. According to WHO statistics, one-third of the world's population is infected with TB. The most serious problem is drug-resistant TB aggravated by the adaptation of new pathogenic strains to weakened immunity.
Prof. Valery Danilenko, the head of the Department of Genetics and Biotechnology at VIGG, comments on the issue: "Humanity is trying to beat the disease with new drugs and innovative treatment methods, but we have—tactically speaking—already lost the battle. During the last 50 years of research, only one antibiotic with a novel type of action has been produced—Bedaquiline. It has been in use for about two years, now. However, mycobacteria have already developed mutations that make them resistant to that drug."
New strains of drug-resistant bacteria with altered virulence have already emerged, exploiting precisely this weakness by targeting immunodeficient patients.
Bioinformatics and genetics help identify a dangerous strain of TB pathogens
Researchers currently identify seven to eight major M. tuberculosis lineages (groups). They differ from one another in mutations of various genes. A genome can have from 300 to 1,000 of such lineage-specific mutations, or SNPs. The term SNP (pronounced "snip") means a mutation in a particular gene involving the substitution of only one nucleotide. If the mutation occurs in a functional part of a virulence gene, the protein encoded by that gene will trigger a different host immune response. This enables the pathogen to overcome host resistance mechanisms developed in childhood as a result of BCG (anti-TB) vaccination.
Natalya Mikheecheva, a researcher at the Laboratory of Bacterial Genetics at VIGG with a bioinformatics degree from MIPT, explains, "We carried out research aimed at identifying the genes and mutations that allow mycobacteria to thrive in people with altered immune status, including HIV-positive patients. We developed a catalog of SNPs in more than 300 virulence genes. Virulence was defined as the ability of a pathogen to cause disease, overcome host resistance via invasion and adhesion to host cells, and adapt to hostile environments, including immune response modulation."
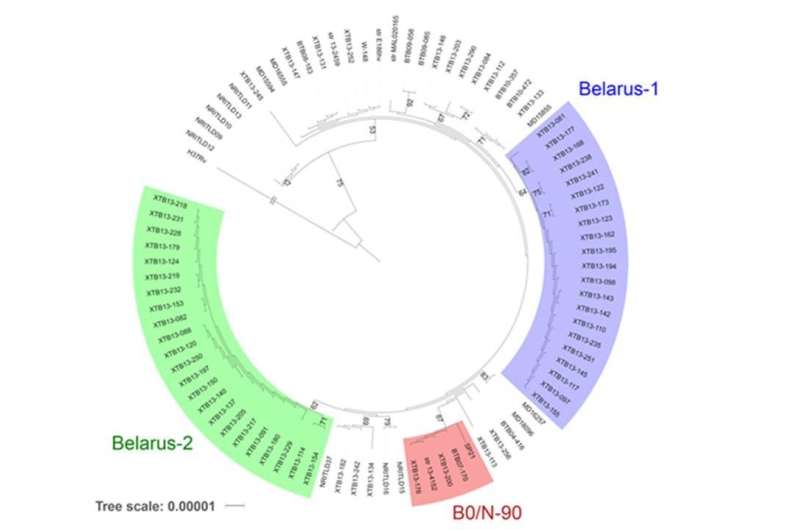
Each lineage was found to comprise dozens or even hundreds of sublineages, depending on the specific gene and the location of the mutation. Bioinformatics analysis conducted using software developed at MIPT's Department of Biological and Medical Physics (MIPT) revealed mutations specific to an epidemiologically dangerous sublineage within the Beijing-B0 lineage.
The scientists used databases of sequenced and described genomes to track the spread of the epidemiologically dangerous B0/N-90 sublineage in Russia and the neighboring European countries Belarus, Moldova and Sweden.
Development of new vaccines
To combat drug-resistant TB, an international consortium called TBResist formed in 2008. Its members include leading experts in medicine, genetics, bioinformatics, etc. from the U.S., Sweden, Russia, the U.K., Bangladesh, Zimbabwe, South Africa, Taiwan, and other countries.
Prof. Danilenko, who led the research in Russia, says, "Our work with the international consortium involved cooperating with our colleagues from South Africa and China to draft a project aimed at investigating the epidemiologically dangerous strain identified in our study. The project is currently being considered by expert communities of the three countries, including the Ministry of Education and Science of the Russian Federation. Our goal is to warn the international community and the health ministries of the BRICS countries of the impending danger. In the '80s, it was HIV. We may well expect something similar from new mutated TB strains—it's a Pandora's box."
Treatments that are available now can cure the disease within a year or two. However, we could see the emergence of mutant pathogens developing rapidly in certain population groups. With the flu, there is an established practice of making a new vaccine every year to counteract the latest mutated strain of influenza. But unlike the influenza virus, which only has several genes, M. tuberculosis has more than 300 virulence genes, each of them potentially subject to mutations.
For the last 30 years, scientists all over the world have been trying to design a genetically engineered TB vaccine. To do this, only certain genes of the bacterium are used, not its whole genome. These genes are cloned to obtain their protein products, which are then used to vaccinate patients and monitor their immune response. There are, however, hundreds of M. tuberculosis sublineages. The research findings indicate that vaccines need to take into account such factors as the host's immune status and the presence in the pathogen of any of at least a dozen epidemiologically dangerous lineages with mutations in particular virulence genes.
Prof. Danilenko drives the point home: "We detected mutations that may enable the bacteria to thrive by exploiting compromised immunity. From that point, it is basically analogous to the flu. We suggest that vaccines against specific TB lineages need to be developed using the genes identified through the bioinformatics analysis of hundreds of sequenced genomes. This will help us to find a basic approach that could inhibit the spread of the dangerous lineages. We have also developed diagnostic tests to identify such lineages."
More information: Natalya E. Mikheecheva et al, A Nonsynonymous SNP Catalog of Mycobacterium tuberculosis Virulence Genes and Its Use for Detecting New Potentially Virulent Sublineages, Genome Biology and Evolution (2017). DOI: 10.1093/gbe/evx053