Study reveals how the most common DNA mutation happens
Shape-shifters aren't just the stuff of fiction, they're real—and they're inside our DNA.
In the Feb. 1 issue of the journal Nature, researchers describe how two normally mismatched bases in human DNA, guanine and thymine, are able to change shape in order to form an inconspicuous rung on the helical DNA "ladder." This allows them to survive by avoiding the body's natural defenses against genetic mutations.
"When these two bases form a hydrogen bond by accident, at first, they don't fit quite right," explained Zucai Suo, professor of chemistry and biochemistry at The Ohio State University and co-corresponding author of the study. "They stick out along the DNA helix, so normally it's easy for the enzymes that replicate DNA to detect them and fix them. But once in a while, before they can be detected, they change shape. It's almost as if the two bases are able to 'fix' each other, so they can fit like a normal base pair and escape DNA repair mechanisms.
"They're bad guys, but they pretend to be good guys to survive," Suo said.
The discovery provides a foundation for work on other types of DNA mutations, which are responsible for diseases as well as normal aging and even evolution.
The four bases of DNA each have their own size and shape, and are supposed to fit together in just the right way. Adenine (A) is always supposed to pair with thymine (T), and cytosine (C) is always supposed to pair with guanine (G). The two "Watson-Crick" base pairs, A-T and C-G, form the DNA sequences of all life as we know it. However, if G were to somehow mispair with T, for example, that would be a mutation.
In fact, the G-T mutation is the single most common mutation in human DNA. It occurs about once in every 10,000 to 100,000 base pairs—which doesn't sound like a lot, until you consider that the human genome contains 3 billion base pairs.
Researchers would like to understand how mutations happen in order to better understand a myriad of diseases, including cancer, that are caused by them. This work provides an important piece of information that researchers can use moving forward in this effort.
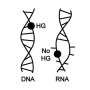
Though scientists had long speculated that the G-T mispair shape-shifted in order to resemble a normal G-C or A-T pair, this phenomenon had not been directly observed until Duke University biochemists, led by Hashim M. Al-Hashimi, used a form of nuclear magnetic resonance imaging to reveal that these Watson-Crick-like G-T mispairs form in so-called "naked" DNA.
Still, the question remained of just how G-T mispairs come to exist. That's why Al-Hashimi contacted Suo at Ohio State, and asked him to help pin down the biochemical mechanism that was responsible.
"An interesting question is: What determines the mutation rate in a living organism," Al-Hashimi said. "From there, we can begin to understand the specific conditions or environmental stressors that can elevate errors."
Suo and doctoral student Walter Zahurancik used a DNA polymerase, an enzyme that replicates DNA, to insert a G-T mispair into a DNA strand. By stopping the chemical reaction at different times and analyzing the resulting DNA molecules, they were able to measure how efficiently the polymerase could form the G-T mispair.
Together, Al-Hashimi and Suo determined that the G and T bases would pair, but in a misshapen way that stuck out from the DNA helix. Then, in a fraction of a second, the bases would re-arrange their chemical bonds so that they could "snap" into the shape of a normal base pair and fool the polymerase into completing the chemical reaction.
In short, they perform a masquerade that enzymes are less likely to detect during DNA replication and repair.
The mutation's survival is a real feat, since it has to overcome a good bit of basic physics. Bases pair in a certain way because of how the protons and electrons in their atoms are arranged. Base pairing requires some amount of energy, and the easiest, most energy-efficient pairs to form are the "right" ones—A-T and C-G.
In effect, the G-T pair has to overcome an energy barrier to form and maintain itself. It turns out that when the G and T bases change shape, they make themselves more energy efficient—still less efficient than a normal base pair, but efficient enough.
Next, the researchers will try to replicate the experiments with another, somewhat less common mutation, the A-C mispair.
More information: Isaac J. Kimsey et al, Dynamic basis for dG•dT misincorporation via tautomerization and ionization, Nature (2018). DOI: 10.1038/nature25487