Molecular doorstop could be key to new tuberculosis drugs
Tuberculosis, which infects roughly one quarter of the world's population and kills nearly two million people a year, is not only deadly but ancient: signs of the disease have been found in Egyptian mummies. Despite its age, Mycobacterium tuberculosis, the pathogen that causes the illness, continues to learn new tricks. It has a particular knack for evolving antibiotic resistance, leaving hundreds of thousands of people with few treatment options.
Now, research conducted by Rockefeller scientists under the direction of Seth Darst, the Jack Fishman Professor, and Elizabeth Campbell, a senior research associate in Darst's laboratory, offers hope for a new and potent weapon against TB. Their work, which appears in eLife, focuses on an antibiotic that kills M. tuberculosis in the laboratory, but is not suitable for clinical use. By explaining how the drug operates, their research might allow others to design new antibiotics that could in fact be used to treat TB patients—and might even work on other bugs.
A Flawed Candidate
Campbell explains that the antibiotic in question, fidaxomicin, is uncommonly adept at killing M. tuberculosis cultivated in the lab. To be useful against TB in the real world, however, an antibiotic must be absorbed by the gut and eventually reach the lungs when taken orally—something that fidaxomicin cannot do.
Fidaxomicin targets an enzyme called RNA polymerase (RNAP), which transcribes DNA into RNA, a process fundamental to life. The enzyme possesses a hinged pincer, or clamp, that swings shut to secure DNA for transcription.
Scientists suspected that the drug works by somehow interfering with this clamp. But they didn't know precisely how the molecule went about its job—knowledge that would be essential to creating more useful versions of fidaxomicin.
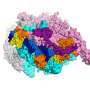
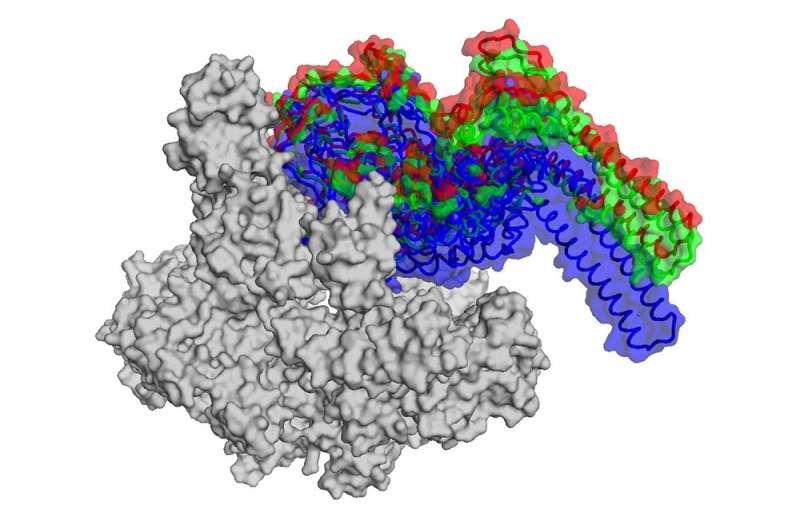
By using a powerful imaging technique known as cryo-electron microscopy, however, Campbell and her colleagues were able to figure out exactly how the antibiotic throws a wrench into RNAP.
Open Wide
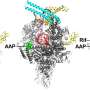
Thanks to earlier research, the team already knew that the version of the enzyme found in M. tuberculosis only works properly when combined with a protein called RbpA—a transcription factor not found in all bacteria. RbpA winds itself into a narrow pocket located at the base of the RNAP clamp, making it fully functional. Using cryo-EM, the researchers were able to show for the first time that fidaxomicin binds to RbpA as well as other parts of the RNAP deep inside that pocket.
What's more, they were able to pinpoint what happens when the two molecules come together: fidaxomicin physically jams the enzyme's clamp open and won't allow it to close.
"It acts like a doorstop, and prevents the clamp from securing DNA for transcription," says Campbell.
Endless Possibilities
Next, Campbell and her associates went a step further. Working with a non-pathogenic cousin of M. tuberculosis called M. smegmatis, the team used a mutant form of the bacteria that lacked the part of RbpA that interacts with fidaxomicin. When exposed to fidaxomicin, normal Mycobacteria could not grow. The mutants, however, were able to thrive despite the presence of the antibiotic and went about multiplying as usual, confirming what the researchers already suspected: that RbpA is an essential part of the mechanism that makes these microbes vulnerable to the drug.
Medicinal chemists might be able to use this insight into how fidaxomicin works to design versions of the antibiotic that are absorbed through the gut, or to identify other drugs that also bind in the RNAP pocket and interact with RpbA. Antibiotics that require RbpA to work would be very useful since they would kill only the select group of bacteria that contain RbpA. Antibiotics that kill indiscriminately can cause significant collateral damage, wiping out benevolent bacteria and breeding resistance among more dangerous ones.
At the same time, because all bacterial RNAP possesses the same pocket that fidaxomicin uses as its binding site, drug developers might also be able to use the team's structural data to develop antibiotics that kill bugs that do not rely upon RbpA at all.
"Our hope is that drug companies will use these studies as a platform for modifying and designing antimicrobials," says Campbell. "They could use the structures we analyzed to design antibiotics that would only inhibit Mycobacteria, but they could probably also design broad-spectrum antibiotics that would kill a wide range of other bacteria."
More information: Hande Boyaci et al. Fidaxomicin jams Mycobacterium tuberculosis RNA polymerase motions needed for initiation via RbpA contacts, eLife (2018). DOI: 10.7554/eLife.34823