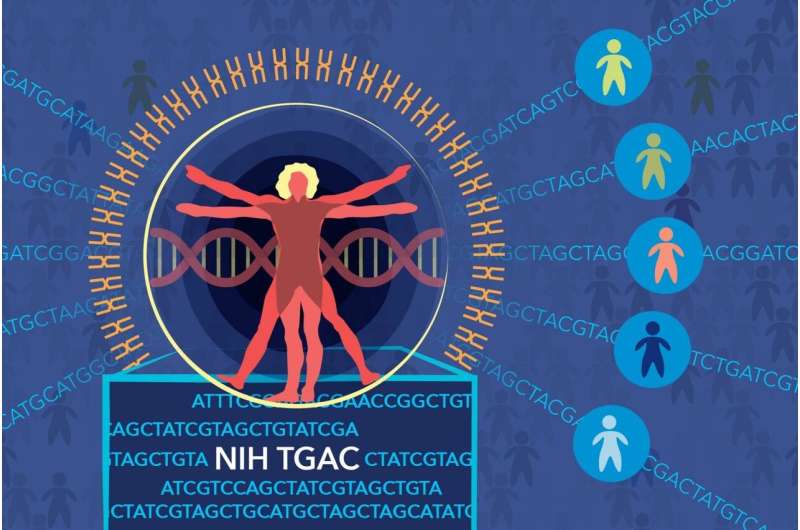
NIH pilot project will match researchers to genes, gene variants of interest
Databases such as the 1000 Genomes Project and the Exome Aggregation Consortium (ExAC) harbor large numbers of genotypes (an individual's collection of genes). Until now, it has been difficult for researchers to re-contact individuals with genotypes of interest and investigate the health consequences of their genes and gene variants.
To address this challenge, National Institutes of Health and Inova Health System researchers are launching The Genomic Ascertainment Cohort (TGAC), a two-year pilot project that will allow them to recall genotyped people and examine the genes and gene variants' influence on their phenotypes, an individual's observable traits, such as height, eye color or blood type.
The project reverses a long-standing research paradigm of examining a person's traits or symptoms and then searching for genes or gene variants that cause or contribute to them. NIH will establish a new database of 10,000 human genomes and exomes, the 1-2 percent of the genome that contains protein-coding genes. Once the database is established, NIH and Inova researchers will predict conditions that specific genes or gene variants might produce and test those predictions by re-examining individuals who donated their DNA sequence information to the database. TGAC will be based in the Washington, D.C., area and only people who've expressly given consent to be re-contacted will participate in TGAC.
"We're trying to advance science in a new, creative and slightly radical way," said Leslie Biesecker, M.D., TGAC co-organizer and chief of the Medical Genomics and Metabolic Genetics Branch at NIH's National Human Genome Research Institute (NHGRI). "Our goal is to determine what genes and gene variants do. We're especially interested in using this as a platform to test our ability to predict phenotype from genotype - one of the key underpinnings of predictive genomic medicine." NHGRI will host the database and administer the program.
Participating NIH institutes will each contribute genome and exome sequences from existing research programs to the database, including NHGRI's CLINSEQ program, healthy bone marrow donors from the National Heart, Lung, and Blood Institute's transplant program, the National Institute of Environmental Health Sciences Environmental Polymorphism Study, and others. Inova will contribute genome sequences of 8,000 people in parent-child trios from its Longitudinal Childhood Genome Study.
"Inova is thrilled to be partnering with NIH on this important work," said John E. Niederhuber, M.D., executive vice president and CEO of Inova Translational Medicine Institute. "When we established the Childhood Longitudinal Cohort study at Inova in 2012, this is exactly the vision we had for how such a longitudinal cohort with genomic data would be used to support ground-breaking genomic research."
An additional 1,000 patients will be recruited to have genome sequencing performed. Because knowledge of how genomic variants and their downstream biological effects vary across populations, half of the newly-recruited patients will be individuals from Hispanic backgrounds. They will complement the 500 individuals with African-American ethnic backgrounds in CLINSEQ.
"Ten thousand genomes and exomes is our initial goal because that will allow recruitment of individuals with both common and rarer gene variants," said Richard Siegel, M.D., Ph.D., TGAC co-organizer, clinical director and chief of the Autoimmunity Branch at NIH's National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS). "This is large-scale sharing of data and research volunteers. We will be taking already sequenced cohorts and making them available to large numbers of researchers."
A researcher might locate a genotype of interest in the database and, if the request is approved, ask participants with the genotype to come to the NIH Clinical Center in Bethesda, Maryland. It is the largest clinical research center in the world with unparalleled phenotyping resources and capabilities. Most of the researchers' requests will require simple blood samples from participants. Other tests might include electroencephalograms (EEG), tests that detect electrical activity in the brain, or MRI scans.
"This is essentially match-making between genes and gene variants and the researchers who study them," said Dr. Siegel. "We hope they'll provide insights that haven't been possible until now. If successful, we plan to open the database to outside researchers in the future."