Developing rapid molecular method for the detection of Plasmodium knowlesi
Malaria is a major public health threat in tropical and subtropical countries. In 2015, an estimated 212 million malaria cases occurred worldwide, resulting in 429000 deaths. Malaysia's aim to eliminate malaria by year 2020 is threatened by the emergence of the zoonotic species Plasmodium knowlesi. It is the predominant malaria species infecting humans in Malaysia and capable of producing severe disease. An early and accurate Point-of-Care (PoC) diagnosis can assist in patient management and reduce mortality.
PoC tests are simple and can be performed at the bedside. To date, PoC diagnosis of malaria is not available in Malaysia and most parts of the world. Researchers from University of Malaya aimed to develop a rapid (< 25 minutes), inexpensive and operationally amenable methodologies derived from recombinase polymerase amplification technique (RPA) in combination with Nucleic acid lateral flow immunoassay for direct visualization detection for PoC diagnosis of malaria. RPA assay have not been implemented for PoC diagnosis. Previous studies shown that malaria detection using dried blood spots (DBS) in nested PCR seemed to be a suitable alternative to microscopy (Smit et.al., 2014). The researchers also believe that DBS will be useful in the RPA-NALFIA test, which can be performed by painless finger-prick blood.
The RPA assay was performed using specifically designed primers and probes. The P. knowlesi small subunit ribosomal RNA (18S rRNA) and Plasmodium genus-specific gene was employed as target gene and the primers and probes were designed. Endpoint detection of the amplification product was performed by lateral flow technology that required a specifically designed lateral flow (LF) probe. The probes consisted of an oligonucleotide backbone with a 5- FAM, a tetrahydrofuran (THF) residue that replaces a nucleotide and a polymerase extension blocking group, C3-spacer at the 3 end.
For detection of Plasmodium species genus, the LF probe was used with a biotin-labelled reverse amplification primer. Another oligonucleotide primer involved in the reaction is a conventional primer. To detect the P. knowlesi genus-specific gene, the LF probe designed is similar to Plasmodium genus gene. However, this probe was used together with digoxigenin-labelled reverse amplification primer at the 5 end.
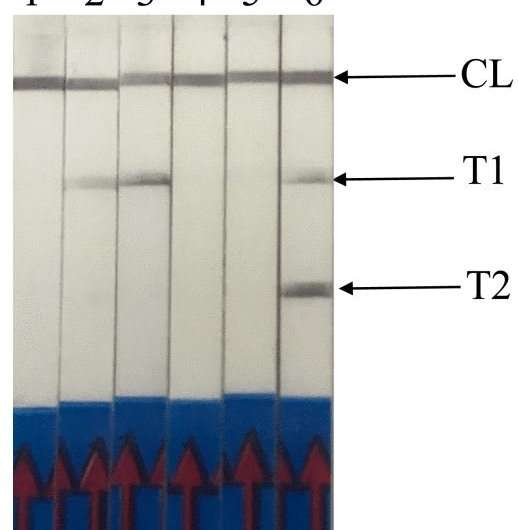
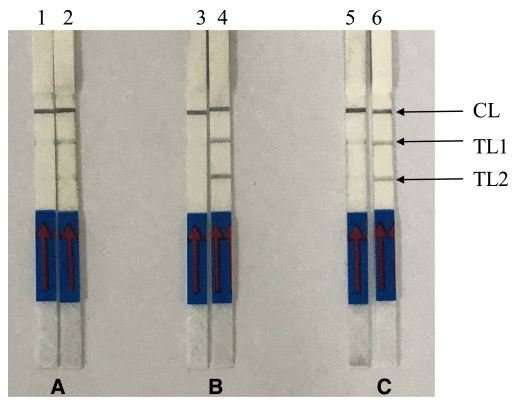
Another oligonucleotide primer involved is a forward amplification primer. RPA assay was conducted using the commercial TwistAmp nfo kit. The reaction tube was then incubated in a heating block with constant temperature at 37 oC for approximately 15 min. For a positive sample, LF-RPA amplicon was observed as two test lines (TL1 and TL2) on the lateral flow strips (Figure 1). TL1 indicates a genus-specific gene and TL2 shows P. knowlesi-specific gene detection. A control line, which is immobilized with anti-rabbit antibodies serves as the RPA assay control.
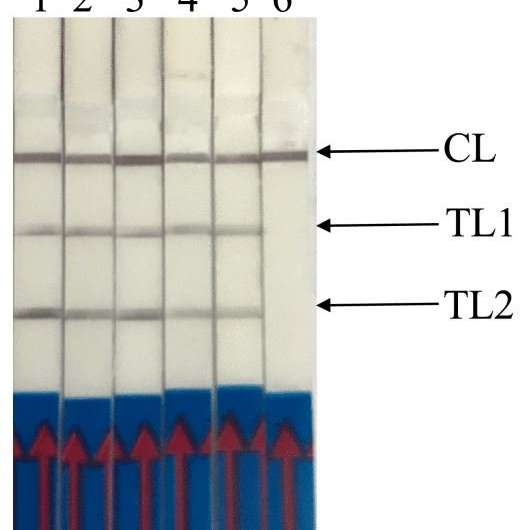
For the evaluation of the signal enhancement, amplified RPA products were tested in different dilution buffers. The researchers found that buffer B (50 mM Trizma-HCl, 150 mM NaCl, 0.05% Tween-20) was the most suitable dilution buffer as the band of amplified RPA product was more visible with less background compared to other dilution buffers. DNeasy Blood and Tissue kit were used to extract and purified the DNA from P. knowlesi strain A1H1 culture for detection limit of the developed LF-RPA assay in this study. The detection limit of P. knowlesi RPA in this work was as low as 10 copies (Figure 3).
The specificity of LF-RPA was tested using DNA templates from other non-P. knowlesi parasites such as Toxoplasma gondii, Sarcocystis sp., P. falciparum, P. vivax and P. ovale. The results shown that LF-RPA was 100% specific for all P. knowlesi samples whereby both test lines and control line were present within 5 min of incubation at room temperature (Figure 1). Meanwhile, only TL1 and control line were present when tested with non-P. knowlesi strains (11 P. falciparum, 12 P. vivax and 2 P. ovale). In further experiments, DNA extracted from eight healthy donors were used as template in the LF-RPA reaction. The LF-RPA did not detect any of the negative DNA samples.
With incubation at 37 oC, the 18S rRNA gene of P. knowlesi was successfully amplified within 12 min. By adding a specifically designed probe to the reaction solution, the amplified RPA product can be visualized on a lateral flow strip. The RPA assay exhibited high sensitivity with limits of detection down to 10 parasites/µl of P. knowlesi. Nonetheless, it was demonstrated that all P. knowlesi (n = 41) and other Plasmodium sp. (n = 25) were positive while negative samples (n = 8) were negative. Therefore, a combination of RPA and lateral flow strip detection is a highly promising approach with the potential to be suitable for use in resource-limited settings.
With the PoC test, detection of malaria infection can be done easily and rapidly in the field or bedside (by finger-prick), thus can rapidly control the outbreaks. The use of the PoC device is applicable to remote area where no lab setup is required to conduct the testing. Only patient detected positive of malaria will be hospitalised for advanced treatment, thus reduces the number of patients under monitoring and unnecessary hospitalization. This testing methodologies has won a Gold medal at the 4th Korea Creative Invention Contest CiC 2017. The researchers plan to develop LF-RPA assay on detection of five human malaria Plasmodium sp (P. knowlesi, P. ovale, P. falciparum, P. vivax and P. malariae) simultaneously. There may be an opportunity to develop the device further for personalised medicine, whereby self-testing is possible even at home without the need of supervision of medical officer.
More information: Meng-Yee Lai et al. Rapid Detection of Plasmodium knowlesi by Isothermal Recombinase Polymerase Amplification Assay, The American Journal of Tropical Medicine and Hygiene (2017). DOI: 10.4269/ajtmh.17-0427
Meng-Yee Lai et al. Recombinase Polymerase Amplification Combined with a Lateral Flow Strip for the Detection of Plasmodium knowlesi, The American Journal of Tropical Medicine and Hygiene (2017). DOI: 10.4269/ajtmh.17-0738