New method for finding disease-susceptibility genes
A new study has resulted in a novel statistical algorithm capable of identifying potential disease genes in a more accurate and cost-effective way. This algorithm is a possible approach for the identification of candidate disease genes, as it works effectively with less genomic data and takes only a minute or two to get results.
This breakthrough has been reported by Professor Dougu Nam and his research team in the School of Life Sciences at UNIST. Their findings were published in Nucleic Acids Research on March 19, 2018.
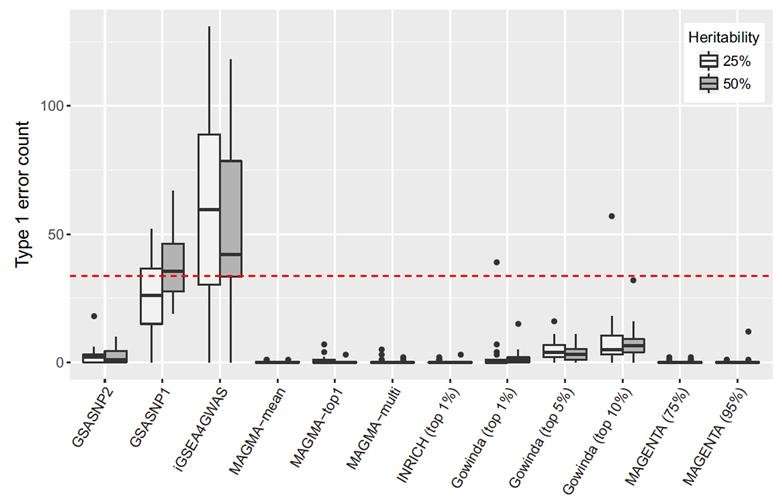
In the study, the research team presented the novel method and software GSA-SNP2 for pathway enrichment analysis of GWAS P-value data. According to the research team, GSA-SNP2 provides high power, decent type I error control and fast computation by incorporating the random set model and SNP-count adjusted gene score.
"GSA-SNP2 is a powerful and efficient tool for pathway enrichment and network analysis of genome-wide association study (GWAS) summary data," says Professor Nam. "With this algorithm, we can easily identify new drug targets, thereby deepening our understanding of diseases, and unlock new therapies to treat it."
Each individual's genome is a unique combination of DNA sequences that play major roles in determining who we are. This accounts for all individual differences, including susceptibility for disease and diverse phenotypes. Such genetic variations among humans are known as single nucleotide polymorphisms (SNPs). SNPs that correlate with specific diseases could serve as predictive biomarkers to aid the development of new drugs. Through the statistical analysis of GWAS summary data, it is possible to identify the disease-associated SNPs.
Despite the astronomical amounts of money and time invested in the statistical analysis of SNP data, the conventional SNP detection technologies have been unable to identify all possible SNPs. This is because most of the conventional methods for detecting SNPs are designed to strictly control false positives in the results. Therefore, among tens of thousands of genomics data and hundreds of thousands of SNPs analyzed, the number of markers described within a candidate disease gene often reaches several tens.
"Although controlling false positive SNPs is needed for the correct interpretation of the results, too much filtering may hamper its usefulness in drug development," says Professor Nam. "Therefore, enhanced statistical power is essential to practical statistical algorithms."
The team aimed to develop an algorithm that improves the statistical predictability while maintaining accurate control of false positives. To do this, they applied the monotone Cubic Spline trend curve to the gene score via the competitive pathway analysis for gene expression data.
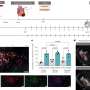
In a comparative study using simulated and real GWAS data, GSA-SNP2 exhibited high power and best prioritized gold standard positive pathways compared with six existing enrichment-based methods and two self-contained methods. Based on these results, the difference between pathway analysis approaches was investigated and the effects of the gene correlation structures on the pathway enrichment analysis were also discussed. In addition, GSA-SNP2 is able to visualize protein interaction networks within and across the significant pathways so that the user can prioritize the core subnetworks for further studies.
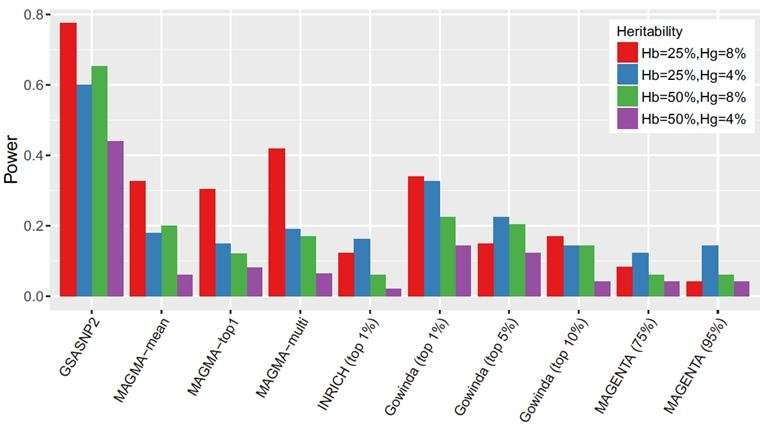
According to the research team, GSA-SNP2 provides a greatly improved type I error control by using the SNP-count adjusted gene scores, while nevertheless preserving high statistical power. It also provides both local and global protein interaction networks in the associated pathways, and may facilitate integrated pathway and network analysis of GWAS data.
The research team expects that their GSA-SNP2 is able to visualize protein interaction networks within and across the significant pathways so that the user can prioritize the core subnetworks for further studies.
More information: Nucleic Acids Research (2018). DOI: 10.1093/nar/gky175