Genome editing helps decipher a congenital liver disease
Congenital hepatic fibrosis (CHF) is a rare genetic disease that causes malformation and fibrosis (scarring) of the liver. Occurring in roughly one out of every 20,000 births, CHF can lead to an enlarged liver, impaired blood flow to the intestines (portal hypertension), infection of the bile ducts and liver failure. In severe cases, a liver transplant is usually needed to treat the disease. In a study published in the Journal of Hepatology, researchers centered at Tokyo Medical and Dental University (TMDU) describe a new experimental model of CHF, which allowed them to uncover molecular players that may contribute to the illness.
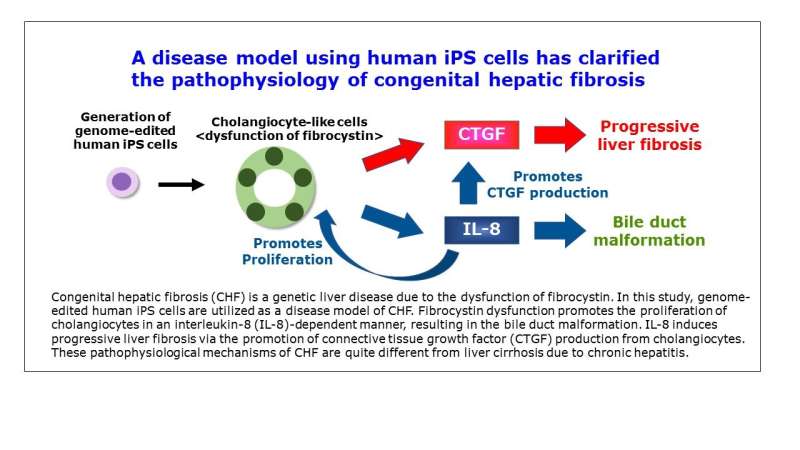
"CHF is actually quite different from more common liver diseases, which typically lead to cirrhosis," corresponding author Sei Kakinuma explains. "For instance, the symptoms of CHF aren't caused by cellular necrosis, as they are in hepatitis. We know that there is an important genetic component underlying CHF, because patients often have mutations in a particular gene, PKHD1, coding fibrocystin. But, it isn't exactly clear how these mutations contribute to the unique symptoms of the disease."
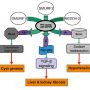
Researchers often rely on experimental models to study CHF—these usually involve mice with mutations in PKHD1. Mouse models, while useful, often have additional liver pathologies that are rarely seen in patients with CHF. Patient-derived stem cells are problematic, too, because mutations in PKDH1 differ from patient to patient, and the impact of different mutations on the disease is poorly understood. To get around these drawbacks, the team used the genome editing technology CRISPR/Cas9, which allowed them to engineer their own human stem cell model of CHF.
"Our approach was to take stem cells from healthy subjects and induce a specific mutation in PKHD1," the first author Tomoyuki Tsunoda explains. "The precision of CRISPR allowed us to make a targeted genetic change that completely abolishes the expression of the protein coded by the gene. We could then assess the molecular and pathological behavior of the stem cells, knowing with confidence that we had rendered the PKHD1 gene completely inactive."
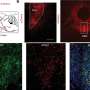
Prior studies had not clearly pinpointed molecules whose expression changed as a result of mutated PKHD1. Equipped with cells whose PKHD1 gene was unambiguously lost, the research team was now able to scour the genetic landscape for key molecular changes. They found that the expression of two genes, IL-8 and CTGF, was greatly increased as a result of the PKHD1 mutation. Both IL-8 and CTGF appear to play critical roles in the progression of the disease and, importantly, were also found to be expressed at abnormally high levels in the livers of CHF patients.
"Our findings suggest that upregulation of IL-8 and CTGF may contribute to inflammation and fibrotic scarring seen in patients with CHF," corresponding author Mamoru Watanabe concludes. "Importantly, we've uncovered a previously unknown pathway that has clear connections to the disease. We think that this discovery could potentially lead to new therapeutic targets to block the progression of CHF, reduce the need for liver transplants, and perhaps even improve the life expectancy of patients."
More information: Tomoyuki Tsunoda et al, Loss of fibrocystin promotes interleukin-8-dependent proliferation and CTGF production of biliary epithelium, Journal of Hepatology (2019). DOI: 10.1016/j.jhep.2019.02.024