Unprecedented exploration generates most comprehensive map of cancer genomes to date

An international team has completed the most comprehensive study of whole cancer genomes to date, significantly improving our fundamental understanding of cancer and signposting new directions for its diagnosis and treatment.
The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Project (PCAWG), known as the Pan-Cancer Project, a collaboration involving more than 1,300 scientists and clinicians from 37 countries, analyzed more than 2,600 genomes of 38 different tumour types, creating a huge resource of primary cancer genomes. This was then the launch-point for 16 working groups studying multiple aspects of cancer's development, causation, progression and classification.
Previous studies focused on the 1 per cent of the genome that codes for proteins, analogous to mapping the coasts of the continents. The Pan-Cancer Project explored in considerably greater detail the remaining 99 per cent of the genome, including key regions that control switching genes on and off—analogous to mapping the interiors of continents versus just their coastlines.
The Pan-Cancer Project has made available a comprehensive resource for cancer genomics research, including the raw genome sequencing data, software for cancer genome analysis, and multiple interactive websites exploring various aspects of the Pan-Cancer Project data.
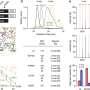
The Pan-Cancer Project extended and advanced methods for analyzing cancer genomes which included cloud computing, and by applying these methods to its large dataset, discovered new knowledge about cancer biology and confirmed important findings of previous studies. In 23 papers published today in Nature and its affiliated journals, the Pan-Cancer Project reports that:
- The cancer genome is finite and knowable, but enormously complicated. By combining sequencing of the whole cancer genome with a suite of analysis tools, we can characterize every genetic change found in a cancer, all the processes that have generated those mutations, and even the order of key events during a cancer's life history.
- Researchers are close to cataloguing all of the biological pathways involved in cancer and having a fuller picture of their actions in the genome. At least one causal mutation was found in virtually all of the cancers analyzed and the processes that generate mutations were found to be hugely diverse—from changes in single DNA letters to the reorganization of whole chromosomes. Multiple novel regions of the genome controlling how genes switch on and off were identified as targets of cancer-causing mutations.
- Through a new method of "carbon dating," Pan-Cancer researchers discovered that it is possible to identify mutations which occurred years, sometimes even decades, before the tumour appears. This opens, theoretically, a window of opportunity for early cancer detection.
- Tumour types can be identified accurately according to the patterns of genetic changes seen throughout the genome, potentially aiding the diagnosis of a patient's cancer where conventional clinical tests could not identify its type. Knowledge of the exact tumour type could also help tailor treatments.
"The findings we have shared with the world today are the culmination of an unparalleled, decade-long collaboration that explored the entire cancer genome," said Dr. Lincoln Stein, member of the Project steering committee and Head of Adaptive Oncology at the Ontario Institute for Cancer Research (OICR). "With the knowledge we have gained about the origins and evolution of tumours, we can develop new tools to detect cancer earlier, develop more targeted therapies and treat patients more successfully."
"This work is helping to answer a long-standing medical difficulty, why two patients with what appear to be the same cancer can have very different outcomes to the same drug treatment. We show that the reasons for these different behaviours are written in the DNA. The genome of each patient's cancer is unique, but there are a finite set of recurring patterns, so with large enough studies we can identify all these patterns to optimize diagnosis and treatment." said Dr. Peter Campbell, member of the Pan-Cancer Project steering committee and Head of Cancer, Ageing and Somatic Mutation at the Wellcome Sanger Institute in the UK.
"This study provides the most complete picture to date of cancer-causing mutations in all parts of the genome. It was a massive team science effort involving researchers spanning the globe," said steering committee member Dr. Josh Stuart, a professor of biomolecular engineering at UC Santa Cruz. "At UC Santa Cruz, our strengths in systems biology and RNA expression helped us connect findings in the previously unexplored noncoding genome with the pathways that lead to cancer. Like a charted map, this new work creates a reference and resource that researchers can use to interpret future data and physicians can use to guide treatment."
"With the continuing drop in sequencing costs and accumulation of genomic data across increasing numbers of patients worldwide, the comprehensive analyses performed in this project will serve as a template for future work and will enable new discoveries in cancer," said steering committee member, Dr. Gad Getz, professor of pathology at the Massachusetts General Hospital and the Broad Institute of MIT and Harvard.
"This huge international study was only possible due to the work and collaboration of more than a thousand researchers and clinicians across the world, and I would like to thank everyone involved," said steering committee member Dr. Jan Korbel from the European Molecular Biology Laboratory (EMBL) in Heidelberg, Germany.
"The completion of this project represents the culmination of more than a decade of ground-breaking work in studying the cancer genome," said Dr. Tom Hudson, Chief Scientific Officer at AbbVie and a founder of the International Cancer Genome Consortium. "When we launched ICGC in 2007, an initiative of this magnitude was unprecedented. I am thrilled that the scientific community has come together to produce this comprehensive study, which enhances our understanding of cancer and fosters the development of new medicines for cancer patients."
"ICGC's latest initiative called ARGO (Accelerating Research in Genomic Oncology) is about the patient, with the goal of delivering to the world 1 million patient-years of precision oncology knowledge to improve human health. This data must be shared across traditional jurisdictional boundaries to realize the full impact of precision medicine, for the benefit of all." said Dr. Andrew Biankin AO, Regius Professor of Surgery and Director of the Wolfson Wohl Cancer Research Centre at the University of Glasgow, and Executive Director, International Cancer Genome Consortium.
"Using the data and infrastructure created by The Cancer Genome Atlas (TCGA) as a blueprint, PCAWG has further improved our understanding of cancer and strengthened our ability to develop successful, international projects of this scale," said Dr. Jean Claude Zenklusen, Ph.D., director of TCGA Program Office at the National Cancer Institute (NCI).
"In addition to benefiting the cancer research field, this collaboration also honors the many patients who donated samples to TCGA—turning their finite gift of tissue into data that can be used infinitely," said Dr. Carolyn Hutter, Ph.D., National Human Genome Research Institute team lead for TCGA.
More information: Nature page: www.nature.com/collections/pcawg/