Scientists uncover structural basis for SARS-CoV-2 inhibition by Remdesivir
A team of Chinese scientists have reported the high-resolution cryo-EM structure of Remdesivir-bound RNA replicase complex from SARS-CoV-2, the infective virus of COVID-19.
The research, published online in Science on May 1, was conducted by Prof. Xu Huaqiang and Prof. Xu Yechun from the Shanghai Institute of Materia Medica (SIMM) of the Chinese Academy of Sciences (CAS), Prof. Zhang Yan from the Zhejiang University School of Medicine, Prof. Zhang Shuyang from Peking Union Medical College and Chinese Academy of Medical Sciences, and their collaborators.
COVID-19 has spread rapidly around the world and is an ongoing humanitarian crisis. Many countries are now facing tremendous challenges in the fight against SARS-CoV-2. Finding an effective treatment is a very urgent matter.
SARS-CoV-2 is a positive-strand RNA virus that mainly infects human cells through the mucosal system. The massive replication of the virus requires the rapid synthesis of its genetic RNA. This process is mediated by a multi-subunit replication transcription complex composed of multiple non-structural proteins (nsp) of the virus. The core element is the replicase complex, which is the core component of coronavirus replication. Numerous nucleoside drugs targeting replicase are currently under clinical testing, including Remdesivir.
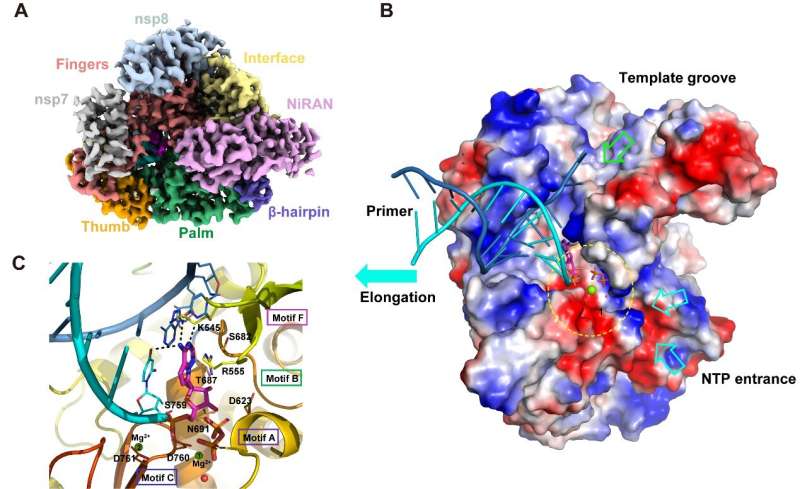
After 46 days of hard work, the scientists were able to elaborate the replicase-targeting mechanism underlying the antiviral efficacy of these nucleoside drugs. Their study reports the cryo-EM structure of the SARS-CoV-2 replicase both in the apo form at 2.8 Å resolution and in complex with a template-primer RNA and Remdesivir at 2.5 Å resolution. The overall conformation of the complex structure is very similar to that of the apo form, with the identical structures at the core catalytic active site. Comprehensive analysis of the structures showed that the SARS-CoV-2 replicase complex is a very efficient enzyme. During RNA extension, conformational change is small, which also explains the highly contagious nature of SARS-CoV-2.
The replicase complex recognizes RNA—but not DNA—through a sequence-independent binding way. The structure of the complex explains how Remdesivir enters the replication active site and covalently links with the viral genome, thereby inhibiting virus replication.
The residues involved in RNA binding as well as those comprising the catalytic active site are highly conserved among most RNA viruses. This shows the conservative mechanism of the replicase complex during gene replication and suggests it may be possible to develop broad spectrum antiviral inhibitors.
The structures described in this study reveal potential binding patterns that offer theoretical support for the design of more powerful, efficient and specific anti-SARS-CoV-2 drugs. In this way, they provide a basis for the design of the antiviral drugs, which are so urgently needed to fight the COVID-19 crisis.
More information: Wanchao Yin et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 01 May 2020: DOI: 10.1126/science.abc1560